Contents
-
Preferences
You can personalize SBTengine with the options in the preferences menu. The following options are available:
1. General
2. GSSP
3. Colors
4. Updates
5. Sample Name / Locus Assignment
6. Loci
7. Library and Data Folders
General
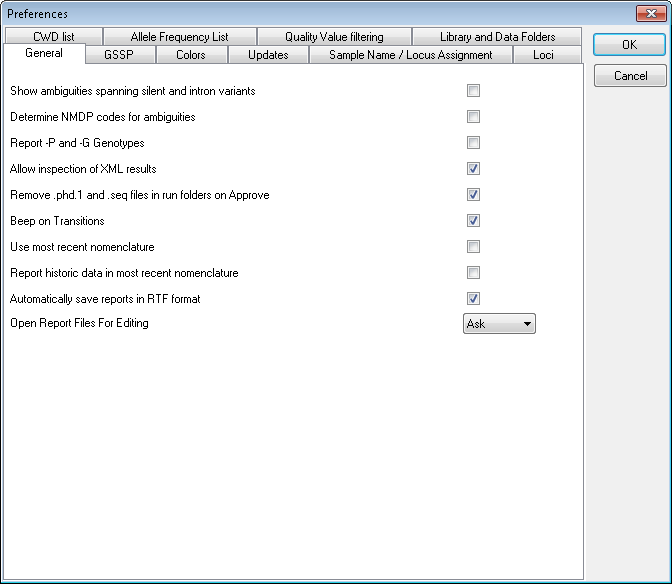
Show ambiguities spanning silent and intron variants
Toggling this tick box shows/ hides ambiguities of alleles that only reflect silent or intron variants. Toggling off is particularly useful in transplantation since these variants are not taken into account for this procedure.
Determine NMDP codes for ambiguities
Allow inspection of XML results
When toggled on you're asked if you want to inspect the xml file after approving the typing of a sample.
Remove *.phd1 files and *.seq files in Run folders on Approve
SBTengine only uses the .ab1 files for analysis. The *.phd1 files are quality score files while *.seq files are text files with the basecalling. In general these are automatically generated by the sequencer for other applications. Ticking this options box will remove these files when a sample is approved, thus keeping your work folder tidy.
Beep on transitions
With this option selected, a beep is played when all inconsistencies in the analysis process are resolved. This way you have an audible aid when the crucial positions are displayed.
Use most recent nomenclature
See New Nomenclature
Report historic data in most recent nomenclature
See New Nomenclature
Automatically save reports in RTF format
Upon approval of a typing, an RTF file of the report is automatically generated and stored in a "Report" folder within the active workfolder.
GSSP
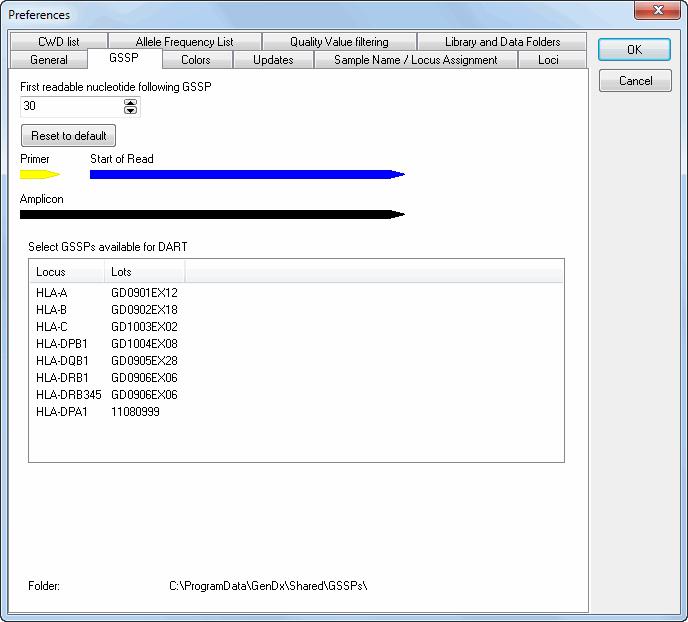
For the selection of the GSSP primers to resolve genotype ambiguities, the DART assumes a default sequence read of 30 basepairs after the end of the primer. Depending on your sequencer and the chemistry this may vary. Should you notice that you have a consistent longer distance between the end of the primer and start of the sequence read you may adjust this in the preferences. This way DART takes this information into account when selecting GSSP primers to resolve the genotype ambiguities.
SBTengine supports the use of different SBTexcellerator GSSP sets. The different sets can be selected based on the SBTexcellerator lot number. Multiple lot numbers can be selected, enabling SBTengine to combine the GSSPs from different lot numbers.
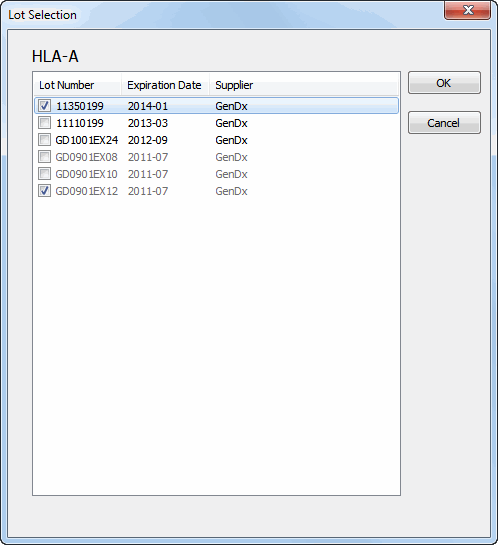
For each locus, lots can be selected. A list of loci is shown with for each locus the lot(s) activated. By clicking on a locus, a list of all available lots with their corresponding expiration date is shown. Lots can be selected and deselected by ticking the checkboxes. After pressing the 'OK' button, these lots are activated.
Colors
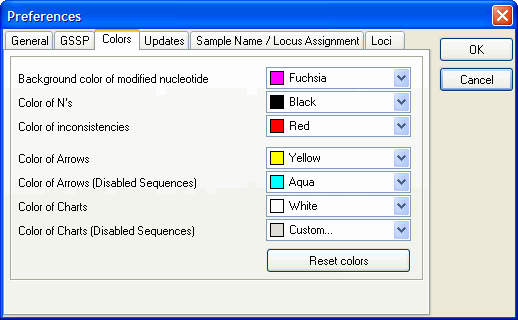
How to adjust preference colors?
In the "File" menu you can find the "Preferences" menu. This menu allows you to set:
the background color of modified nucleotides
color of N's
color of inconsistencies
color of arrows
color of arrows (Disabled Sequences)
color of charts
color of charts (Disabled Sequences)
by clicking the drop-down menu and selecting your desired color.
By clicking the "Reset colors" button, the default colors can be reset.
Updates
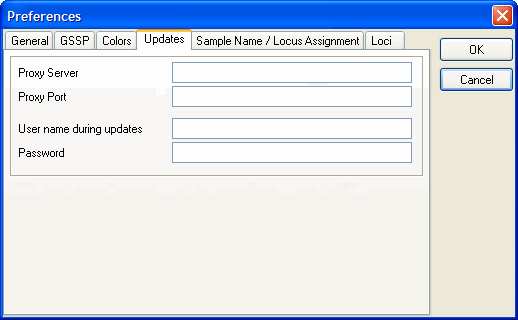
If you are not running SBTengine as an "administrator" you most likely will have limited rights and will therefore not be able to fully complete automatic updating. To circumvent this inconvenience the administrator may fill in his username and password in the appropriate fields within "Preferences". Now updates will be carried out successfully as only this process is carried out as an "administrator. This is an elegant way of securing accessibility to the computer as requested by the general administrator, without limiting the profound advantages of automatic updates via the internet.
User and password Information of the administrator will be securely stored and is not accessible to others.
The network of your institute may limit your access to the internet. In this case the automatic updates may not work. Your IT specialist may enter the Proxy Server and the Proxy Port to facilitate accessibility of SBTengine updates. However, these fields are empty by default and should only be completed when requested by authorized GenDx personnel.
Sample Name / Locus Assignment
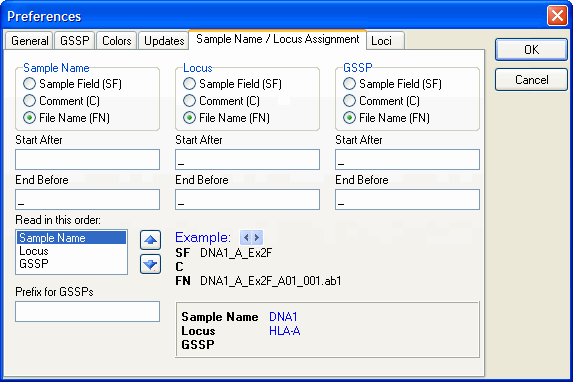
You can set the automic naming for the sample name and locus with this option. Read the Automatic Name / Locus Assignment for more detailed information
Loci
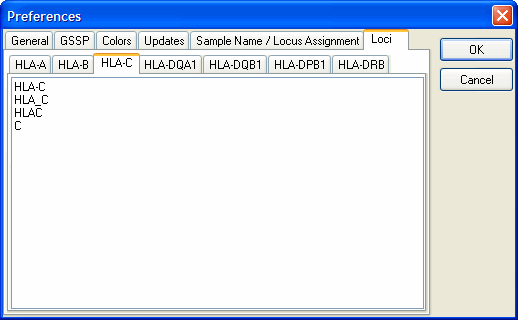
With this option you can set the text for the loci by which SBTengine assigns the locus. For example, the locus HLA-C is assigned to sequence files where the locus is indicated by the text:
HLA-C
HLA_C
HLAC
C
You can also define your own text by adding it to the list.
Library and Data Folders
The "Library and Data Folders" tab allows you to change the data folder locations used by SBTengine. This can be useful if you wish to use a central shared network location such that all users use the same libraries and data files at the same time or if you are using custom data.
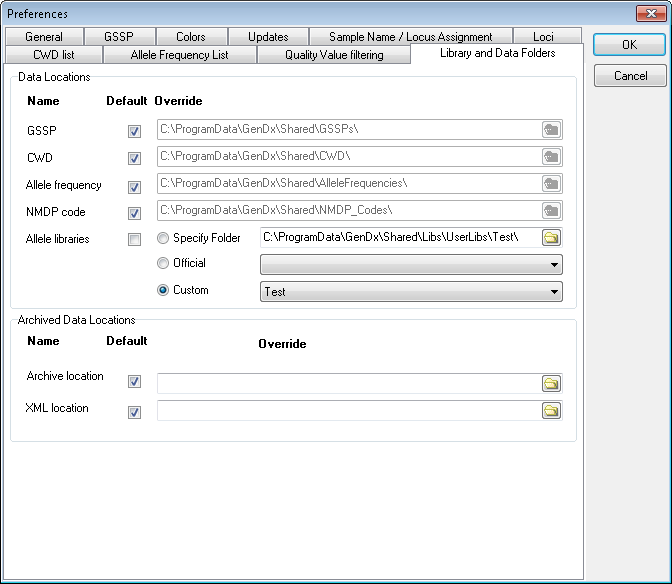
All data files installed with SBTengine are stored at folders in a folder referred to as GenDxRoot. The exact path of GenDxRoot depends on your version and installation of windows. The default locations are shown below, assuming Windows is installed on your C drive:
Windows XP 'C:\Documents and Settings\All Users\Application Data\GenDx\'
Windows Vista 'C:\ProgramData\GenDx\'
Windows 7 'C:\ProgramData\GenDx\'
Each data location can be set to the default setting by checking the corresponding checkbox in the default column, which selects the data installed with SBTengine. Unchecking the default column for a folder location unlocks the edit box to its right, allowing you to specify a custom location for each type of data individually. The data files at the default location are automatically updates when you update SBTengine. Default settings are recommended.
CWD lists
CWD lists can be used to organize the exact matching genotypes. In addition, they can be used to select the crucial positions. GenDx ships a CWD list which is based on the original publication of Cano P. et al: "Human Immunology 68, 392-417 (2007). You can design your own 'CWD-like' list. If placed in GenDxRoot\Shared\CWD\, this file can be selected through the CWD List Preferences.
Note: one can add a file, but can not modify the original file installed with SBTengine. If one wants to make changes to this file, first make a copy and perform the changes in this file.
For information on how to modify or make a CWD list file yourself, please refer to this section.
Allele frequency lists
Allele frequency lists can be used to sort the exact matching genotypes on expected frequency. In addition, they can be used to sort the crucial positions. GenDx ships a number of allele frequency tables based on data published by the NMDP. Details about the source of the data can be found in the header of the files. The default path where Allele Frequency lists are expected is GenDxRoot\Shared\ AlleleFrequencies\. This path can be changed through 'File' 'Preferences' , 'Library and Data Folders'.
Notice, one can add a file, but can not modify the original file shipped with SBTengine. If one wants to make changes to this file, first make a copy and perform the changes in this file. The active Allele Frequency list can be selected through the 'Select Allele Frequency list' dropdown box at 'File' 'Preferences', 'Allele Frequency List'.
For information on how to modify or make a frequency file yourself, please refer to this section.
How choose an alternative archive folder?
Checking the checkbox in the default column results in the archive folder being the "Archive" folder in (relative to) your work folder. In the box to the right you can manually specify an Archive folder that does not have to be in your work folder. Keep in mind that the manually specified archive folder stays the same if you open another work folder.
Note: Only alter these settings after intructions of GenDx personnel!
How to redefine the location of the XML folder?
Checking the checkbox in the default column results in the XML folder being the "XML" folder in (relative to) your archive folder. In the box to the right you can manually specify an XML folder that does not have to be in your archive folder.
Note: Only alter these settings after intructions of GenDx personnel!
How to redefine the location of the allele library folder?
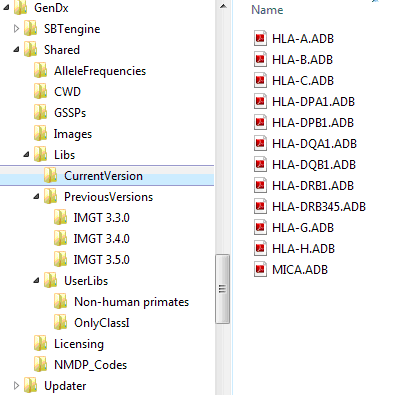
The default allele library locations is GenDxRoot\Shared\Libs\
At this location, three folders are present:
'CurrenVersion': Contains the allele libraries of the current version, installed by SBTengine
'PreviousVersions': Contains allele libraries of previous versions, each in separate folder, installed by SBTengine.
'UserLibs': Can be used by the user to maintain user-defined allele library combinations,
You can select 4 sources for allele libraries:
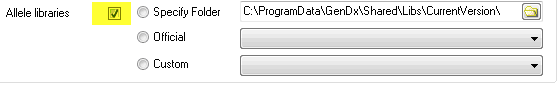
1) Check the default checkbox. This selects the allele libraries installed with SBTengine.
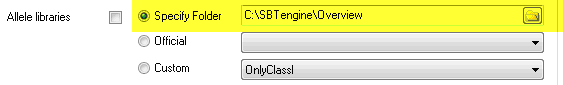
2) Select 'Specify Folder' and browse to a folder containing allele library files.
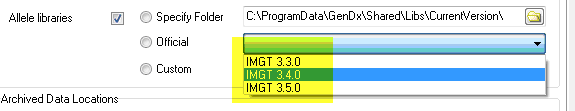
3) Select 'Official' and select the library you need from the pulldown menu next to 'Official'. This option is reserved for libraries previously released by GenDx.
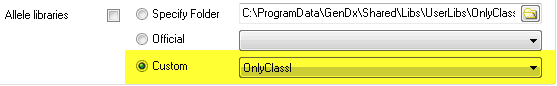
4) 'Select 'Custom' and select a custom allele libraries folder:
a. First, create in a folder in GenDxRoot\Shared\Libs\UserLibs\
b. Copy the libraries you need into this folder, e.g. only the class I libraries of a particular release
c. Reopen the preferences screen in SBTengine
d. Select the folder you just created from the pulldown menu next to 'Custom'.
Source #1: Default libraries
Source #2: Library files in user-defined folder
Source #3: Older library versions
Source #4: Custom user data
An example of a folder structure is shown below: