Contents -
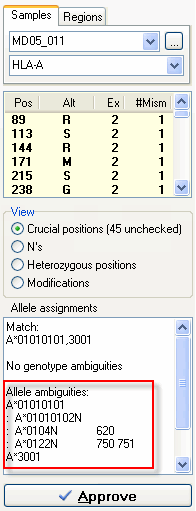 For this sample we got at match HLA-A*01010101,3001. However, the A*01010101 allele is ambigious with the A*01010102N, A*0104N, A*0122N alleles. DART indicates that the A*01010101 allele differs from the A*0104N at position 620 and differs from A*0122N at positions 750 and 751. In the exon overview at the top of the Sequence Overview window you can see that these positions are located in exon 4. By sequencing exon 4 of this sample and reanalyzing the data you resolve these allele ambiguities.
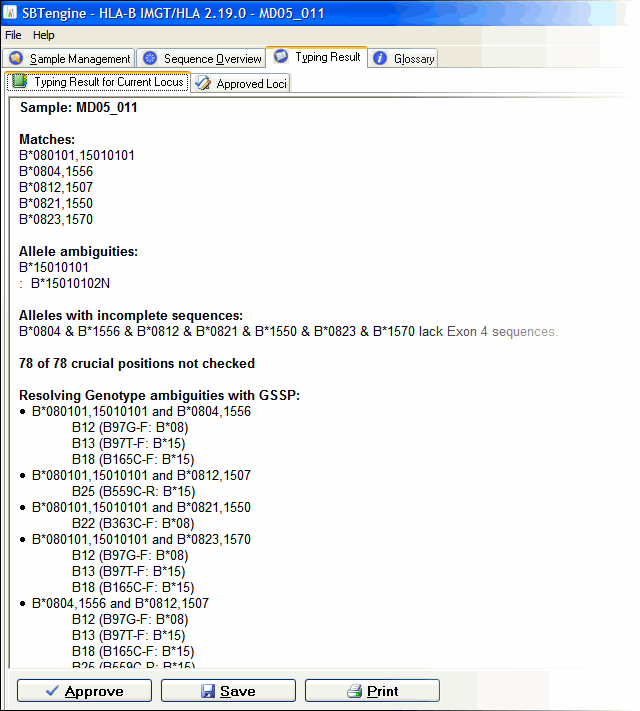
For this sample we got at match HLA-A*01010101,3001. However, the A*01010101 allele is ambigious with the A*01010102N, A*0104N, A*0122N alleles. DART indicates that the A*01010101 allele differs from the A*0104N at position 620 and differs from A*0122N at positions 750 and 751. In the exon overview at the top of the Sequence Overview window you can see that these positions are located in exon 4. By sequencing exon 4 of this sample and reanalyzing the data you resolve these allele ambiguities.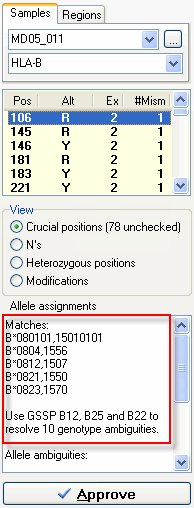 For this sample we have multiple genotypes (B*080101,15010101, B*0804,1556, B*0812,1507, B*0821,1550, B*0823,1570) that match the heterozygous sequence data. To determine which one is the correct one we use GSSPs to seperate the individual alleles. DART indicates which GSSPs to use from the SBTexcellerator kits to resolve these ambiguties, in this case GSSP B12, B25 and B22. Just sequence the stored PCR product with these GSSPs and reanalyze the data in SBTengine. The ambiguities will be resolved.
For this sample we have multiple genotypes (B*080101,15010101, B*0804,1556, B*0812,1507, B*0821,1550, B*0823,1570) that match the heterozygous sequence data. To determine which one is the correct one we use GSSPs to seperate the individual alleles. DART indicates which GSSPs to use from the SBTexcellerator kits to resolve these ambiguties, in this case GSSP B12, B25 and B22. Just sequence the stored PCR product with these GSSPs and reanalyze the data in SBTengine. The ambiguities will be resolved.