Contents
-
Ambiguities
So what are we talking about when we mention the term ambiguities? When the HLA sequences of a DNA sample matches with several allele combinations, we call this an ambigious typing (e.a. an ambiguity). This means that a number of HLA allele combinations can not be excluded based on the sequence information that was obtained. There are three kinds of ambiguities:
Alleles differ only in regions where the sequence of some other alleles are unknown.
Allele Ambiguities
Alleles differ outside the region that is sequenced.
Genotype Ambiguities
Several heterozygous allele combinations (genotypes) give identical heterozygous sequences.
An example is shown below.
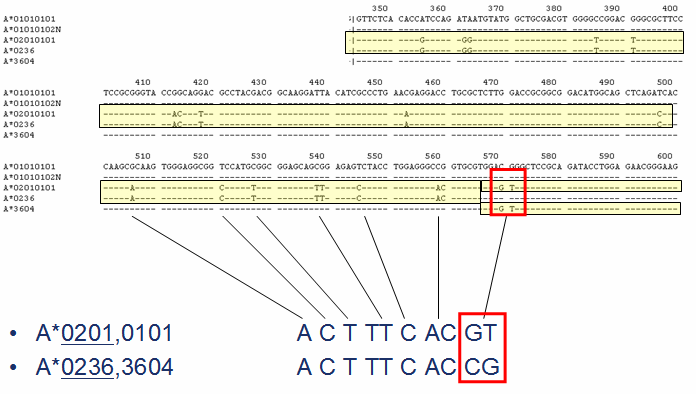
Ambiguities can be resolved in varies ways. For example, allele ambiguities can be resolved by simply including (sequencing) the additional regions that are required to distinguish the allele combinations in the analysis.
Genotype ambiguities can be resolved by:
Group Specific Sequence Primers (GSSP). This requires additional sequencing reactions.
Group Specific Amplification Primers (GSAP). This requires additional amplification and sequencing.
SSP
SSOP
The SBTexcellerator reagents line for high resolution HLA typing solely uses GSSPs to resolve genotype ambiguities. With this approach, you only need to focus on the sequencing technique. Together with the DART technology of the SBTengine software package which indicates what GSSP primer to use, you have a convenient and easy way to fully resolve ambiguities.
