Contents
-
CWD List
The CWD list is a list of Common and Well Documented HLA alleles. SBTengine can use CWD lists to:
1. indicate in the allele assignment which alleles are present in the CWD list.
Genotypes are indicated as:
cwd-,- both alleles are not on the CWD list
cwd+,- the first allele is present on the CWD list but the second is not
cwd-,+ the first allele is not present on the CWD list but the second allele is
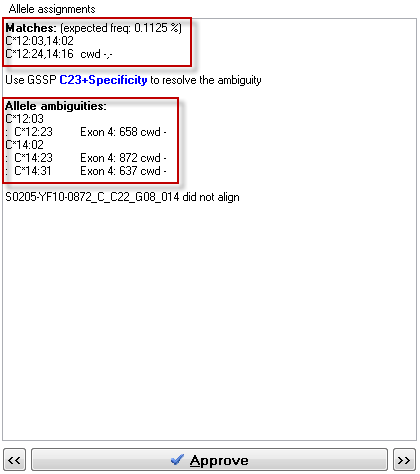
When both alleles of the genotype are on the CWD list, nothing is indicated (i.e. cwd+,+ is not shown). When multiple genotypes match the sequence data, the genotypes are sorted from cwd+,+ to cwd-,-.
Alleles are indicated as:
cwd+ the allele is present on the CWD list
cwd- the allele is not present on the CWD list
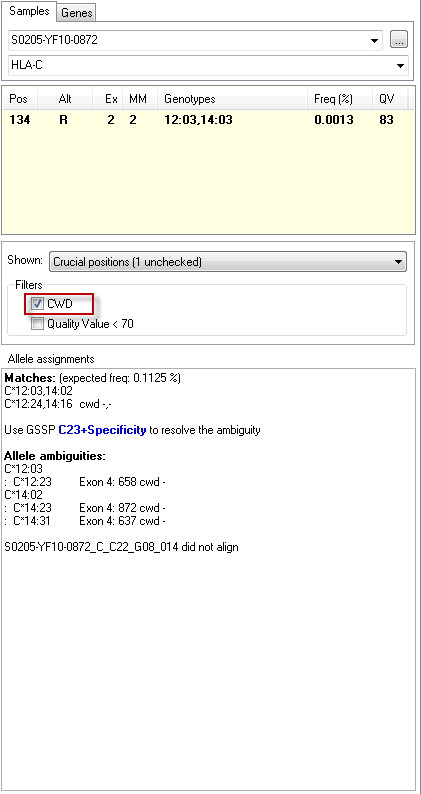
2. filter the crucial postions for genotypes of which both alleles are present in the CWD list. Activating the check box "CWD" filter (see image below) will show only crucial positions of near-matching genotypes with both alleles on the CWD list. This reduces the number of crucial positions to be checked. Please take note that this does NOT exclude allele assignment of genotypes that are not on the CWD list.
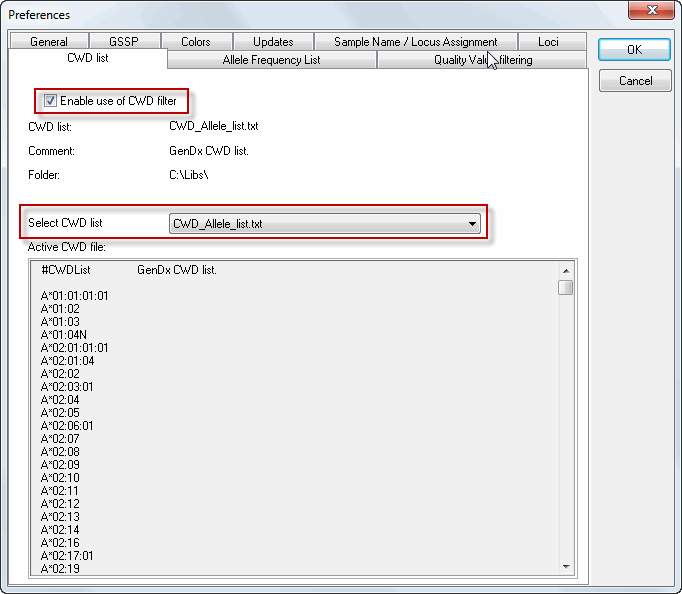
The CWD list functionality is standardly set to 'off'. This can be verified by the greyed out CWD filter check box. One can activate the CWD functionality and select a CWD list from the "CWD List" tab in the preferences (Goto File, Preferences, CWD List tab).
CWD list file format.
A CWD list is a text file with the following format:
First line: #CWDList<[Tab]comment>
Comment lines start with #d
For each locus:
Line [locusname]
Lines allelename
An example:
#CWDList List of Common and Well-documented HLA Alleles.
#d Some comment
[HLA-A]
A*01:01:01:01
A*01:02
A*01:03
A*01:04N
[HLA-B]
B*07:02:01
B*07:02:04
B*07:03
B*07:04
B*07:05:01


