Contents
-
Allele Frequencies
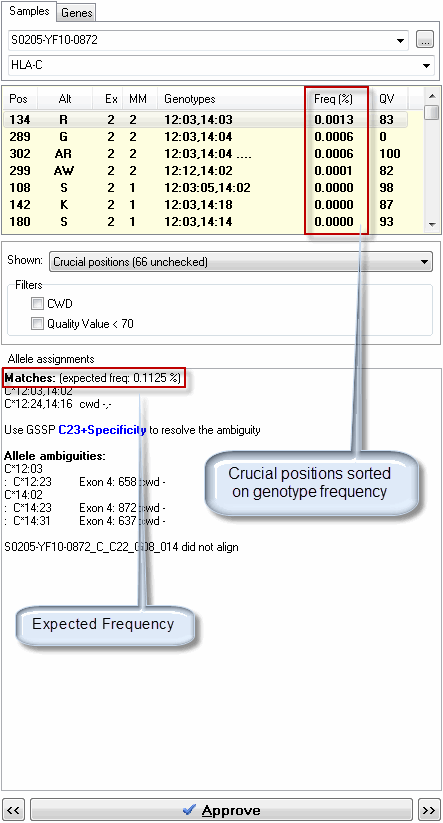
SBTengine can use allele frequency information to calculate the genotype frequency. This illustrates the likelihood of occurence of the genotype in a specific population. Sorting the crucial postions based on the genotype frequencies is very helpful in analysis.
Advantages
The most frequently occuring near-matching genotypes will be listed first. This is advantageous in the following situations:
1. when no match is found, the most likely genotype is listed on top of the crucial positions list irrespective of the number of mismatches. In most cases this will directly point you to the final match.
2. when a match is found, the most likely near-match is listed on top. This is the one which is most relevant for further inspection.
SBTengine will also calculate the expected frequency of the "Matches" (i.e. the sum of all genotype and allele ambiguity frequencies) in the "Allele Assignments" (see image below).
Activation
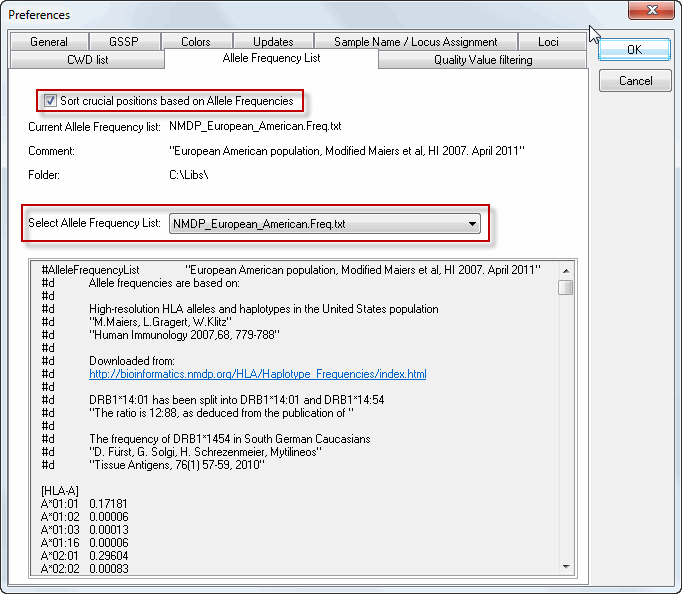
One can activate the sorting of crucial positions based on Allele Frequencies (by clicking the check box) and can select an Allele Frequency List (from the drop down list) in the "Allele Frequency List" tab in the Preferences menu.
Customizing the allele frequency list
GenDx delivers a number of allele frequency lists. You are free to customize these lists specifically to your needs.
However, it is important to maintain the format of the lists.
#AlleleFrequencyList<TAB>"Your Comment"
#d<TAB>any comment by the user
#d<TAB>as many lines as needed
[locusName1]
AlleleName1<TAB>Frequency
AlleleName2<TAB>Frequency
AlleleNameN<TAB>Frequency
[LocusName2]
Please note: <TAB> is a TAB character!
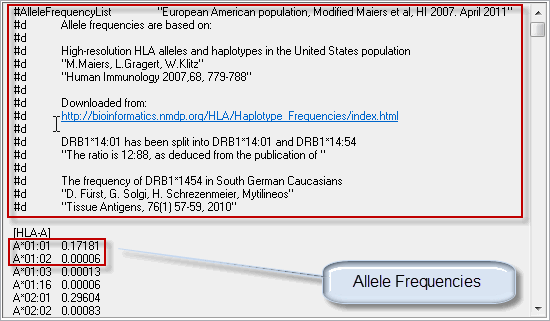
The source of the allele frequency data is described at the top of the list. This information is preceded by "#d" on each line. When you customize the allele frequency list to your needs, you can enter specific descriptive information here (unlimited number of lines).
Allele frequencies are listed in the table as shown in the image below. Alleles of one locus should be alleles that are not defined in the list and are considered to have a frequency of 0%. In the example below one can see that A*01:01 is set to 0.17181 (~17.2%). This entails that the A*01:01:01:01 (first allele name matching A*01:01) is 17.2% whereas the A*01:01:02 is 0%.
SBTengine normalizes the frequencies to a total of 100%. Therefore one may use frequencies as well as allele counts in the list.
When you have contradicting data on the allele frequencies in your population (e.g. A*01:01:01:01 is 10% and A*01:01:02 is 7.2%), you can customize the allele frequency list. Change A*01:01:01 to 10% and add A*01:01:02 with 7.2% to the list.
The allele frequency lists are stored in the Libs folder (typically C:\Program Files\GenDx\SBTengine\Libs) as *.Freq.txt files and can be edited by any word processor. We recommend you first make a copy of the allele frequency list file, rename it and then start editting. The new file should be stored in the Libs folder. If the edited file is not renamed it will be overwritten by the original file after each SBTengine update!




