Operating procedure
General
The user is requested to properly document each sequencing run within the operating software of the automatic DNA-sequencer as supplied by the manufacturer. To facilitate the analysis procedure we advise you to name each sequence file according to an uniform format. For example, start with the sample name (e.g. DNA1), use an underscore ( _ ) as a separator, followed by the locus name (e.g. HLA-C), another underscore and end with the exon and strand that is sequenced (e.g. exon 5 forward strand). This would result in the following file name: DNA1_HLA-C_Ex5F
We advise you to store the sequencing data in a dedicated SBTengine analysis folder (e.g. SBTdata). If your computers are connected through a network we recommend to store the sequence data on a file server that is accessible from each workstation where SBTengine is installed.
If the computers are not part of a network and hence are stand-alone computers we recommend to use one computer for SBT analysis with SBTengine. You can generate a dedicated 'SBTdata' folder on the hard drive. Please make sure that you regularly make backups form this computer to prevent accidental loss of data.
Whether you use a networked environment or not, in this manual we will refer to the folder containing the sequencing files as the 'SBTdata' folder.
The SBTdata folder contains all run folders from the sequencer. SBTengine needs to know the location of the SBTdata folder. You only need to make this setting once per installation of SBTengine.
Instructions
1. When the sequencing run is completed you need to copy the complete run folder to the SBTdata folder.
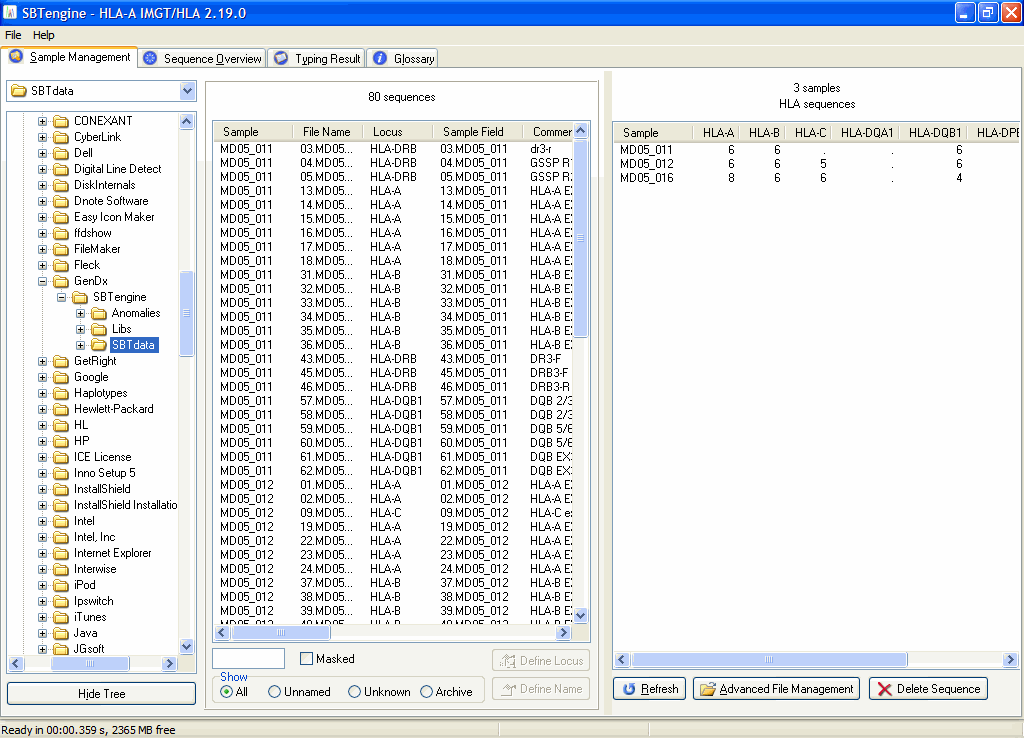
2. Start SBTengine, go to the Sample Management window and select the SBTdata folder in the left pane.
3. Click the 'Hide Tree' button if you want to zoom in on all sequences in the specified SBTdata folder and hide the folder tree. This is particularly useful to prevent accidental deselection of your SBTdata folder.
All sequence files in the run folders that were copied to the SBTdata folder will appear in the middle pane. SBTengine registers all sequence files in the SBTdata folder and all sub folders. Sequence files located in a deeper level of the SBTdata folder are not detected.
An overview of the sequence files is given in the right pane: the number of ABI sequence files per HLA locus is shown for each sample name. Below the right pane you can click the 'Refresh' button to refresh the Sample Management. This is particularly useful when new run folders were added while SBTengine was running. The list is automatically refreshed with each start up of SBTengine. Using the "Advanced File Management" button you may access the sequence files in the Windows Explorer. By clicking the "Delete Sequence" button you delete the selected sequence files shown in the middle pane. This can be useful to remove sequence files of empty (water containing) wells in your sequencer plates.
Follow one of the two procedures below to assign a sample and locus name to each sequence file: