Typing Result for Current Locus
Typing Result for Current Locus
In the Typing Result for Current Locus, the sample name, matches, and ambiguities are described for the current locus, i.e. the sample that is loaded in the Sequence Overview window. GSSPs resolving the ambiguous genotypes are suggested. The Typing Result for Current Locus tab can help you find alternative strategies to resolve genotype ambiguities using GSSPs. Please take note that this is NOT a report for approved samples
See the example below:
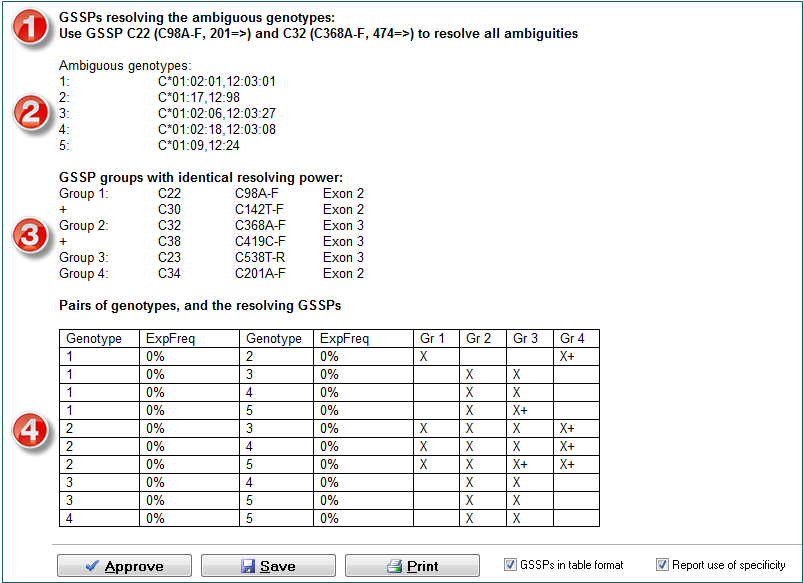
(1) DART reports that GSSPs C22 and C32 are the best solution to resolve all genotype ambiguities.
(2) All ambiguous genotypes are shown. In this case there are 5 ambiguous genotypes.
(3) All GSSPs that can resolve the ambiguities are shown. GSSPs in the same group have identical resolving power, so C22 and C30 resolve the same ambiguity.
(4) Each genotype is paired with another in order to show for each ambiguity which group of GSSPs can resolve the ambiguity. For example, in the first row the ambiguity of genotype 1 and 2 can be resolved by using a GSSP either from group 1 or from group 4. This means C22, C30, or C34 will resolve this ambiguity. From the table it can be seen that almost all ambiguities can be resolved by using a GSSP from group 2 or 3, except for the first one, which needs a GSSP from group 1 or 4. To resolve all ambiguities, the best option is to use a GSSP from group 1 and a GSSP from group 2. This is why DART reports that C22 and C32 should be used. Note that some GSSPs in group 3 and 4 have a + sign. This means that specificity has to be used with these GSSPs. These GSSPs can be ignored by unchecking the "Report use of specificity" checkbox.
If you prefer to see the results in a list instead of a table, the "GSSPs in table format" checkbox can be unchecked:
