Ambiguities and DART
Ambiguities and DART
Ambiguities
When the HLA sequences of a DNA sample matches with several allele combinations, we call this an ambigious typing (an ambiguity). This means that a number of HLA allele combinations can not be excluded based on the sequence information that was obtained. There are three kinds of ambiguities:
- Alleles differ only in regions where the sequence of some other alleles are unknown.
- Allele Ambiguities
Alleles differ outside the region that is sequenced.
- Genotype Ambiguities
Several heterozygous allele combinations (genotypes) give identical heterozygous sequences.
An example is shown below.
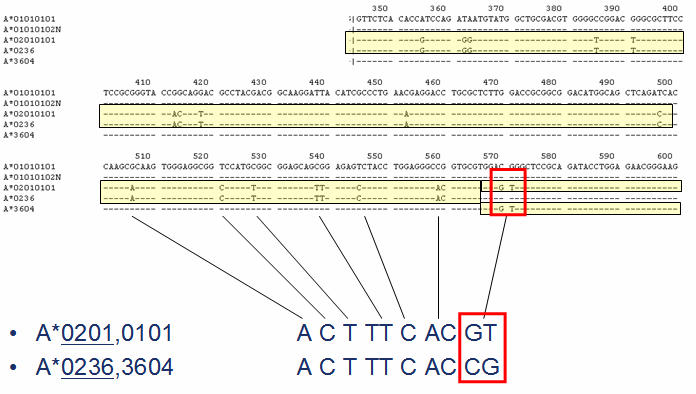
Ambiguities can be resolved in varies ways. For example, allele ambiguities can be resolved by simply including (sequencing) the additional regions that are required to distinguish the allele combinations in the analysis.
Genotype ambiguities can be resolved by:
- Group Specific Sequence Primers (GSSP). This requires additional sequencing reactions.
- Group Specific Amplification Primers (GSAP). This requires additional amplification and sequencing.
- SSP
- SSOP
The SBTexcellerator reagents line for high resolution HLA typing solely uses GSSPs to resolve genotype ambiguities. With this approach, you only need to focus on the sequencing technique. Together with the DART technology of the SBTengine software package which indicates what GSSP primer to use, you have a convenient and easy way to fully resolve ambiguities.
DART
The Dynamic Ambiguity Resolving Tool (DART) is an integrated module in SBTengine, which has been specifically designed to help resolve ambiguities.
When you have established an allele assignment, you may end up with ambiguities in ~ 50% of your samples. DART indicates what sequence information is required to resolve the ambiguities.
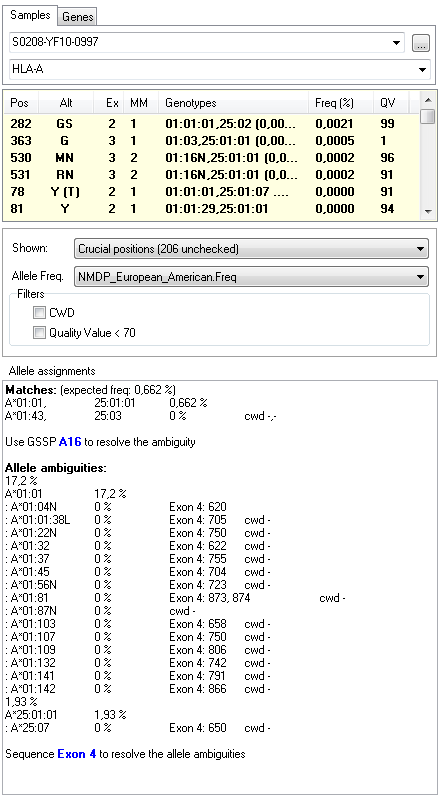
Genotype Ambiguities
For the sample shown above, we have two genotypes (A*01:01, 25:01:01 and A*01:43, 25:03) that both match the heterozygous sequence data. To determine which one is the correct one we use GSSPs to separate the individual alleles. DART indicates which GSSPs to use from the SBTexcellerator kits to resolve these ambiguities, in this case GSSP A16. You can just sequence the stored PCR product with this GSSP and reanalyze the data in SBTengine to resolve all genotype ambiguities.
Allele Ambiguities
Allele ambiguities are reported under 'allele ambiguities'. For the sample shown above, we have a match A*01:01, 25:01:01. However, the A*01:01 allele is ambiguous with 15 other alleles. Similarly, the A*25:01:01 allele is ambiguous with one other allele (A*25:07). DART indicates at which positions these ambiguous alleles differ from each other and reports that by sequencing exon 4 you will be able to resolve these ambiguities.