Typing Result Approved loci
Typing Result for Approved loci
Under the Typing Result tab you can find the tab "Approved loci". Here you can find the information on typing results. These reports can be printed directly or saved as Rich Text File (*.rtf) by clicking the Print or Save button respectively at the bottom of the window.
When you save the report as a *.rtf file, you can open it in any text editor (e.g. Microsoft Word) and add your letterhead and additional text to create a report for the clinician. You may even consider to generate a macro in your text editor to automate this. Please refer to your text editor software for details on how to generate macro's.
Current sample and All samples
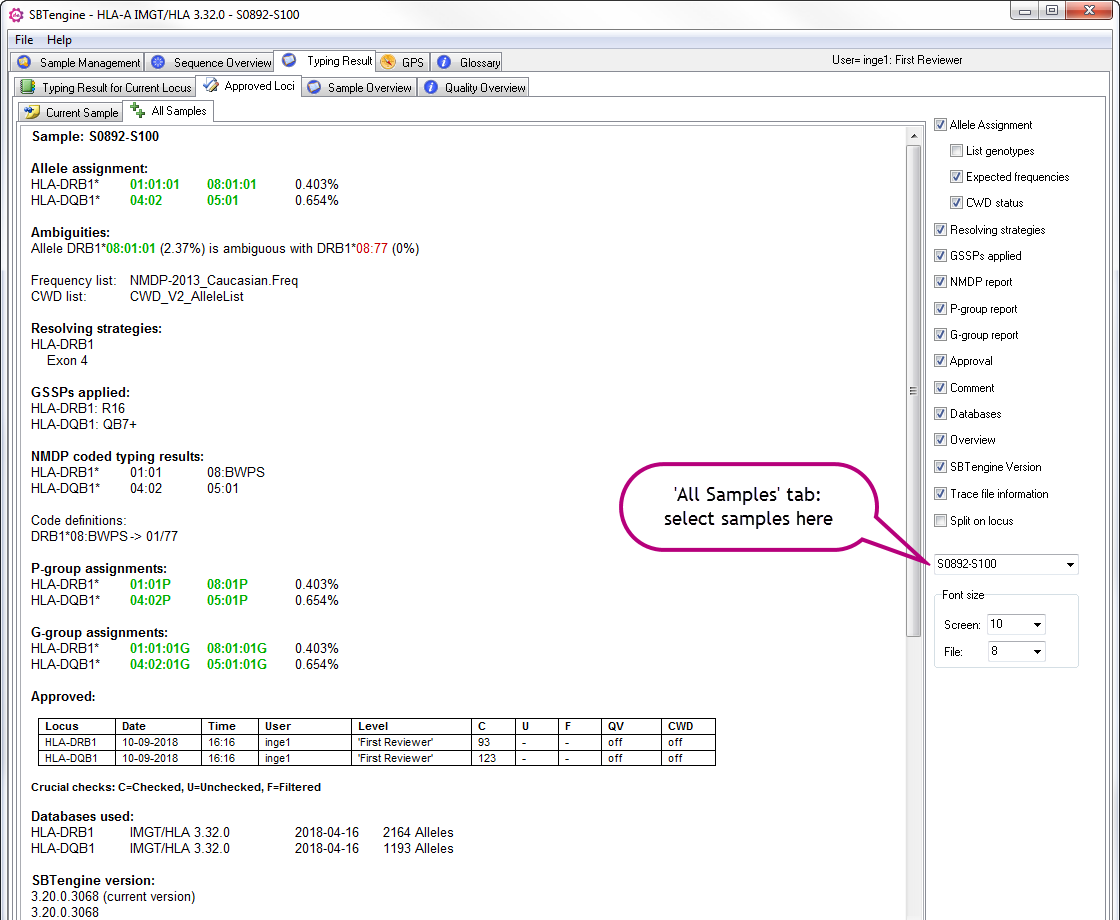
Under the Approved loci tab you can find report information on the Current Sample and on All Samples by selecting the relevant tab. Whereas the Current Sample tab shows the report for the sample that was currently analyzed in the Sequence Overview, the All Samples tab gives you access to the report of all previously analyzed samples. On the right side of the window you can select a sample in the drop down menu.
In both the Current Sample and All Samples tab you have the possibilty to toggle on and off the following diaplay options:
- Allele Assignment: overview of the HLA allele assignment for each approved locus
- List genotypes: ambiguities will be included in Allele assignment list instead of a separate ambiguities list
- Expected frequencies: frequencies are shown
- CWD status: alleles are colored green (CWD) or red (not CWD)
- Resolving strategies: strategies that can be applied to resolve remaining ambiguities
- GSSPs applied: GSSPs that have been used
- NMDP report: NMDP coded typing results
- P-group report: P-group assignments
- G-group report: G-group assignments
- Approval: overview who approved the allele assignment, at what date and time, for each approved locus. Each appoval action is stored.
- Comment: comment that was typed upon approval
- Databases: an overview of which IMGT/HLA database was used for the allele assignment, including the number of alleles, for each approved locus
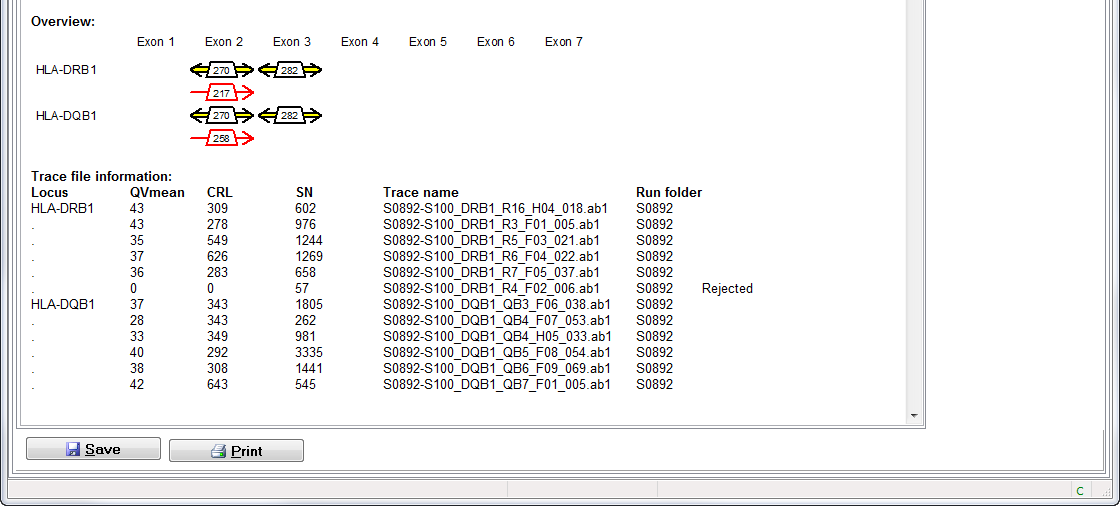
- Overview: this overview shows for each locus what sequence information was used for the allele assigment. A forward and reversed arrow indicate a forward and reverse sequence respectively. The yellow color indicates that the sequence was heterozygous, whereas the red color indicates a homozygous sequence.
- SBTengine version: the version used
- Trace file information: names and QVs of all sequences
- Split on locus: a separate report will be generated for each locus