Submission tool
The submission tool can be used to create files which are needed for the submission of a new allele to the NCBI GenBank and subsequently to the IMGT database.
The submission of alleles is only possible if for class I exon 2 + intron 2 + exon 3, and for class II exon 2 is fully covered and phased.
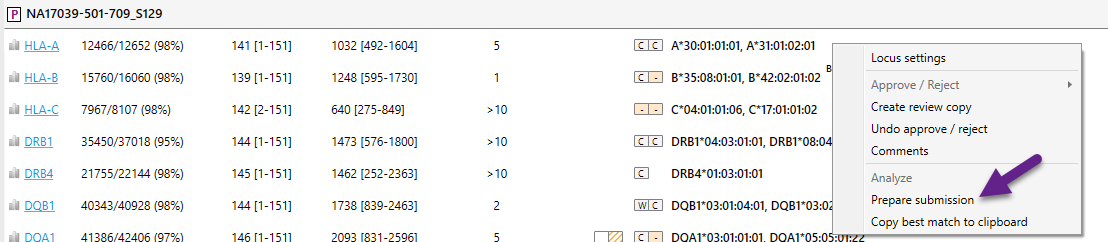
To create the files, right click with your mouse behind the locus which has a new allele and select 'Prepare submission':
When this has been done a message will appear that NGSengine has created submission files in a new folder called 'Submission' in the 'Project' folder:
The submission files can now be found in the 'Submission' folder:
In this folder, 4 separate files can be found for this locus. For each allele of this locus a Fasta file (.fsa) and a Table file (.tbl) have been created:
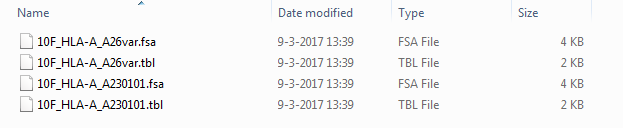
The file names can have the following format:
- [Sample]_[Locus]_[Allelename]var in case the sequence contains exon mismatches (example above: 10F_HLA-A_A26var).
- [Sample]_[Locus]_[Allelename] in case the sequence contains no new information (example above: 10F_HLA-A_A230101). In most cases this is the second allele for the locus which contains a new allele.
- [Sample]_[Locus]_[Allelename]extended in case the sequence contains an allele extension (example: 10F_HLA-B_B51extended). This means that the sequence contains data which is not known in the IMGT database.
The .fsa file contains the sequence of the largest phased region containing the Core+ region of the allele, from beginning to end, in Fasta format. To make this sequence, NGSengine starts with the Core+ sequence and then extends this sequence to the left and to the right until the phasing stops.
The .tbl file contains a feature table with information about the location of the exons and introns, in GenBank format.
Please contact us for more information on how to submit an allele to GenBank.