Single base edit
To make a single base edit, right click on one of the positions in the phasing track:
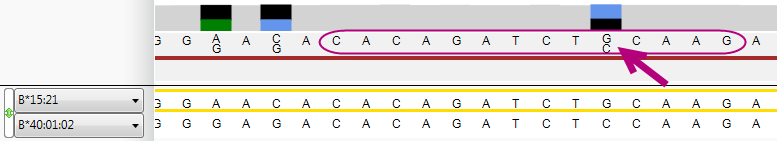
A pop-up window will appear that shows the options for this position and the coverage statistics, with the most frequent base shown on top:
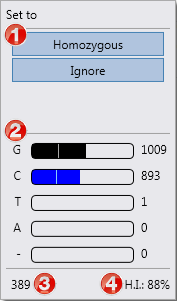
1. Options for this position (see below).
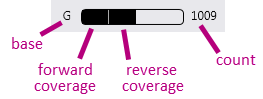
2. Coverage statistics per base. For each base the counts are shown.
3. The gDNA number of the position.
4. The heterozygosity index (H.I.) for the position. The second most occurring base divided by the most occurring base on a position, shown in percentages. A H.I. of 100% is a perfect heterozygous position, a H.I. of 0% is a perfect homozygous position.
In the coverage statistics per base the following information is shown:
The options per position are:
- Homozygous: Mark this position as homozygous, instead of heterozygous. The most frequent base will be used.
- Heterozygous: Mark this position as heterozygous, instead of homozygous. This is only available when a second base is present at this position.
- Ignore: This position will be ignored during analysis when this edit is applied.
- Undo: Only available for a pending edit. Right clicking the same base again will show an Undo option. When this option is used it will immediatley remove the pending edit.
- Reset: Only available for an applied edit. Right clicking the same base again will show a Reset option. When this option is used, it will be shown as a pending edit and will only be saved when it has been applied.
The position where the edit has been made is made visible in the Alignment screen. The position is made purple and the asterisk or pencil are shown:
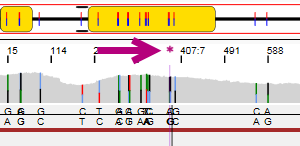
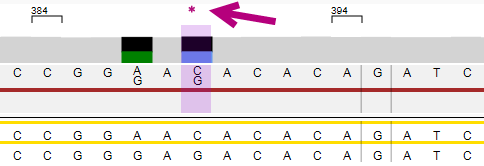
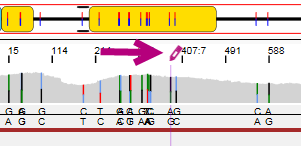
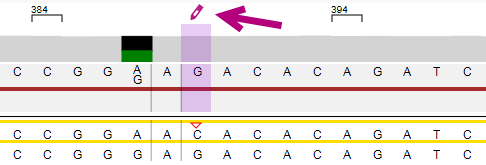
The arrows at the bottom left of the analysis screen can be used to jump to the next (pending) position or phasing area that was manually edited.
![]()