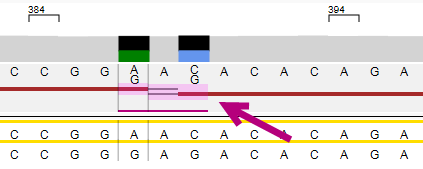
Phasing edit
To make a phasing edit, right click on or below the phasing line at the position:
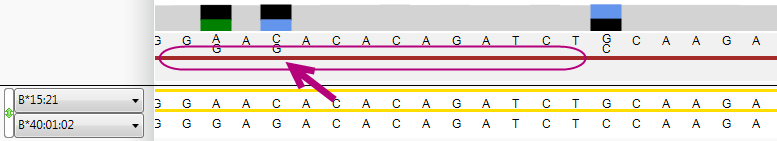
A pop-up window will appear that shows the options for this position, and the statistics of the two positions and their underlying phasing:
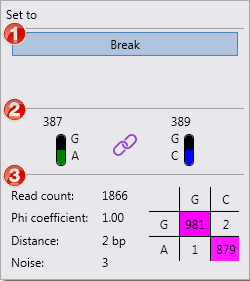
1. Options for this position.
2. The gDNA number of the heterozygous positions and the two most frequent bases at these positions. The symbol in the middle shows whether the positions are linked or unlinked .
3. Statistics:
- Read count: coverage of both heterozygous positions. In the table on the right the counts are shown for the combination of the bases: in this example, a G at position 387 coupled to a G at position 389 was found in 981 reads, a G at position 387 coupled to a C at position 389 was found in 2 reads, etc. The combinations with the most counts are highlighted in pink.
- Phi coefficient: statistical value which indicates how strong the link is, on a scale of 0 (no link) to 1.00 (strong link).
- Distance: the distance in base pairs between the heterozygous positions.
- Noise: the read counts not covered by the phasing table.
The options per position are:
- Break: This will break the phasing region between the heterozygous positions shown in the pop-up window.
- Join: The heterozygous positions shown in the pop-up window will be considered for phasing.
- Undo break: Only available for a pending edit. Right clicking the same position again will show an Undo break option. When this option is used it will immediately remove the pending edit.
- Reset: Only available for an applied edit. Right clicking the same position again will show a Reset option. When this option is used, it will be shown as a pending edit and will only be saved when it has been applied.
The position where the phasing edit has been made is made visible in the Alignment screen.
After "Break" has been selected and is a pending edit, the phasing line between the positions is shown as a dashed line:
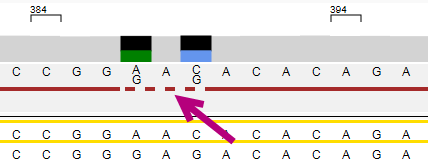
After the phasing edit has been applied, the phasing line is broken and the edit is shown in pink with a purple line below: