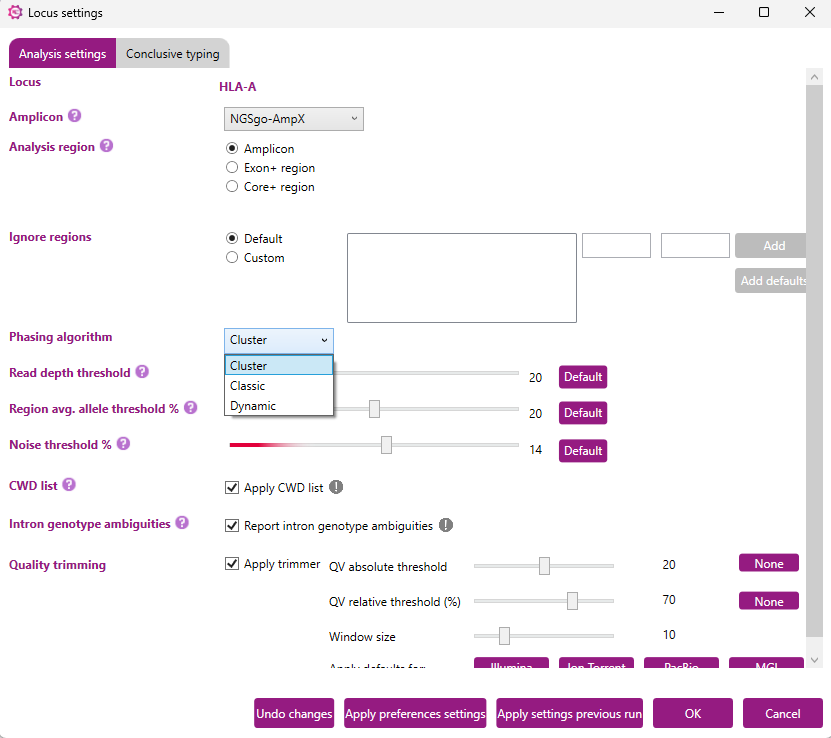
Phasing algorithm
NGSengine® is equipped with three independent Phasing algorithms: the Classic, Cluster, and Dynamic algorithms.
Classic
Based on statistical analysis of all the reads, the Classic algorithm determines for each position whether it is homozygous or heterozygous. When heterozygous, the Classic algorithm determines for each heterozygous position its phase compared to the neighboring heterozygous positions.
Cluster
The Cluster phasing algorithm separates the reads into two groups by identifying groups of reads that are likely to originate from the same allele.
Dynamic
The Dynamic phasing algorithm separates the reads by identifying clusters of reads that are likely to originate from the same allele. Potential overlap between different clusters is taken along in the analysis. In addition, this algorithm incorporates the basecall QV values and can detect local allele imbalances.
The Dynamic phasing algorithm is recommended as the default setting. Overall, this algorithm results in the best performance.
The phasing algorithm can be changed via the Locus default settings in the Preferences menu: