Quality Report
Quality Report
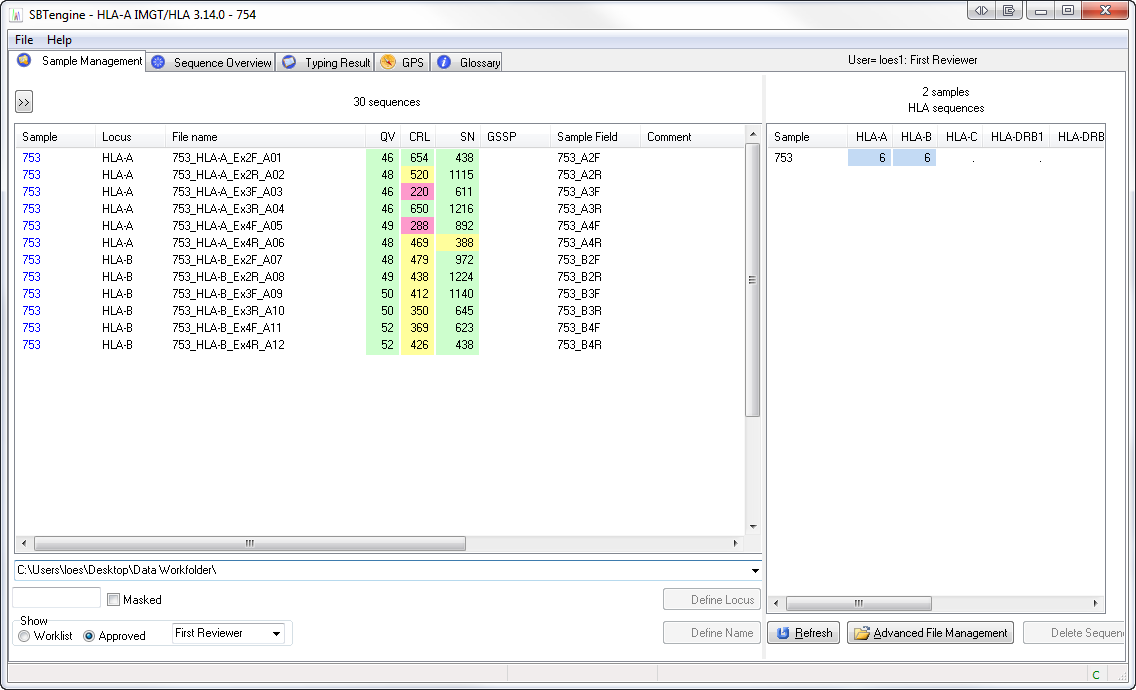
SBTengine displays quality values (QVs) for each sequence trace displayed in the Sample Management window.
- The sequence quality of each sequence trace can be derived from three values: the CRL, QV and S/N%.
- The color patterns applied can be used to obtain an overview of the quality of sequence traces. The CRL, QV and S/N% values of each trace are shown in the Sample Management window. High, intermediate and low QV, CRL and S/N% values are displayed in the colors green, yellow and red, respectively. The color thresholds can be adjusted by changing the CV, CRL and S/N% threshold values in the preference settings.
- CRL (Contiguous Read Length):
The CRL is the longest uninterrupted stretch of bases with a good quality value.
The default threshold setting between low and intermediate is [CRL300] and between intermediate and high is [CRL600].
- QV (Quality Value):
The QV in the Sequence Management window is the average base call quality in the CRL.
The default threshold setting between low and intermediate is [QV20] and between intermediate and high is [QV30].
- S/N% (Signal Noise Ratio):
S/N% is the mean value of the G, A, T and C S/N% as indicated in the trace file.
The default threshold setting between low and intermediate is [S/N40] and between intermediate and high is [S/N400].
This setting applies for the Genetic Analyzer ABI 3730. For the ABI3130 the following settings can be used: [S/N20] and [S/N200].
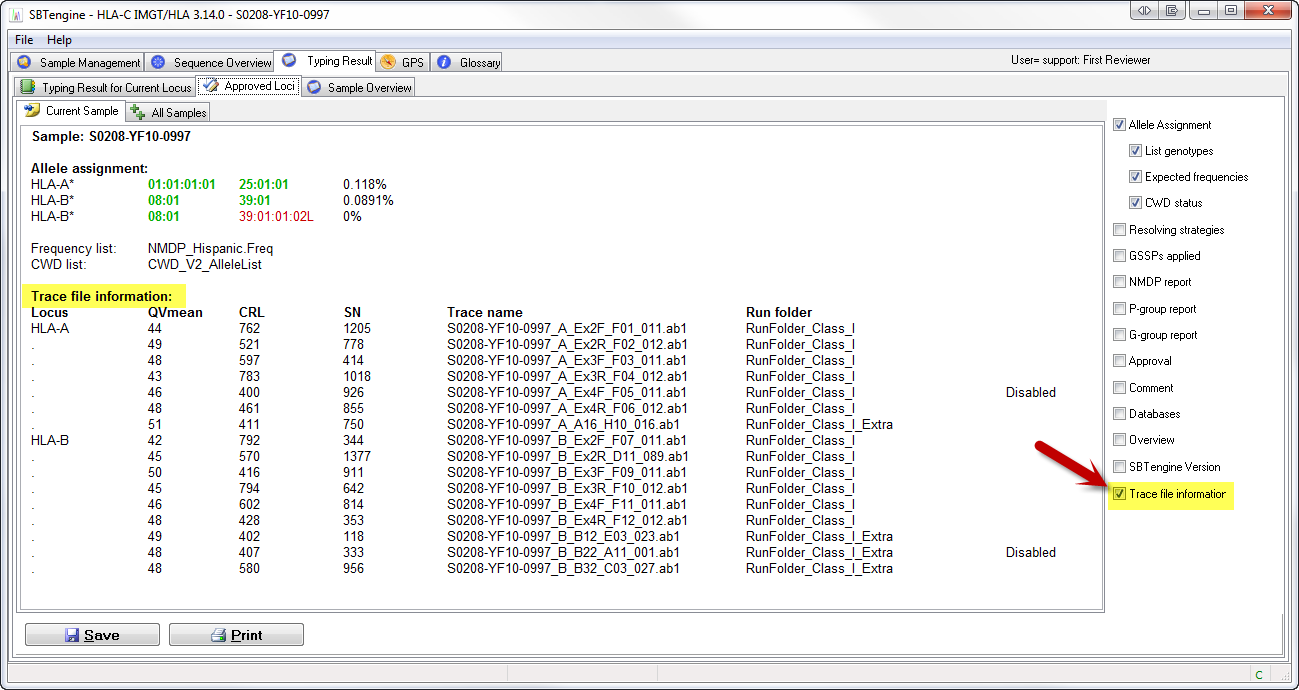
Quality report in the typing result window
After the approval of a sample, you can generate a quality report based on the CRL, QV and S/N% values. The quality report is included in the typing report of that sample by checking the box 'Trace file information'.
The typing report gives the QV, CRL and S/N% values for all the sequence traces of a sample. The report also indicates which trace files have been used for the analysis. Traces that are excluded from analysis are indicated with the comment 'disabled' or 'rejected'.