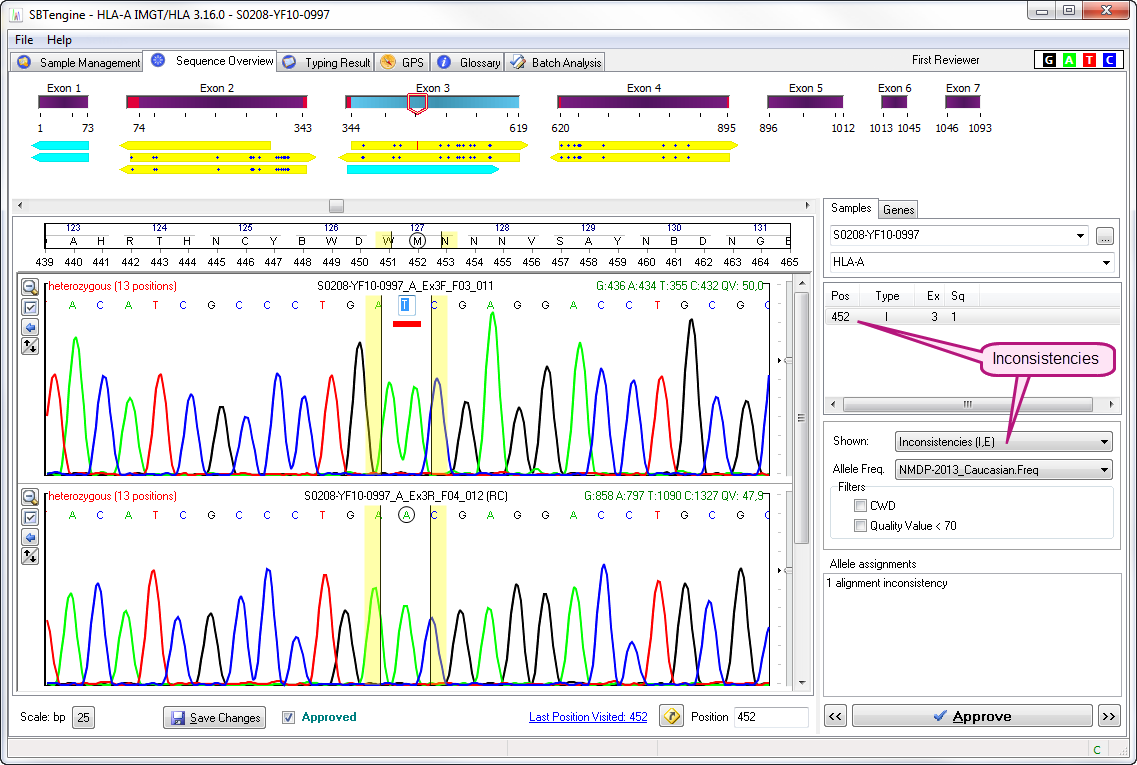
Inconsistent positions
Inconsistent positions
Inconsistencies are specific positions in the sample sequence that do not align with the IMGT/HLA database after merging of the sample sequence files with the IMGT/HLA database. These inconsistencies often reflect false basecalling by SBTengine due to poor sequence quality at these positions. Inconsistencies need to be addressed before you can continue analysis. SBTengine will indicate and list the inconsistencies in the window on the right hand of the electropherograms. The position, type and exon where the inconsistency is located are shown.
Inconsistency types:
I: Alignment inconsistencies
E: Merging Inconsistencies
Position dependent inconsistencies:
E1: Discrepancies between heterozygous sequences
E2: Positions where more than 2 different nucleotides are identified
E4: Positions where homozygous sequences have a ucleotide more than the heterozygote
E8: Positions where the hterozygote has more nucleotide than the homozygous sequences
Inconsistencies can be edited. After pressing ENTER, the next inconsistency is shown. When you have solved the inconsistencies, SBTengine will show the list of crucial positions.