Contents
-
Data Analysis
Analysis of the sequence files is performed in the Sequence Overview window in three main steps:
Step 1
You select the sample and HLA locus that you want to analyze. All ABI sequence files of the selected sample are loaded and aligned with the IMGT/HLA database. In case previously analyzed and approved sequences of the selected sample are present in the Archive folder, these will be imported too.

Step 2
When there are any inconsistencies between the sequences and the IMGT/HLA database, SBTengine will indicate these in the window on the right hand of the electropherograms. The inconsistencies need to be addressed before you can continue analysis. These inconsistencies are indicated by a horizontal red bar below the basecalling in the electropherogram and as a vertical red bar in the yellow sequence arrows. The first inconsistency is shown in a list in the right pane and can be edited. After you have changed the base calling, press ENTER. SBTengine will automatically jump to the next inconsistency. Continue in this fashion until the list with inconsistencies is empty. When finished, SBTengine will report "Alignment OK" in the lower right corner.
Subsequently, inconsistencies between the sequence files (electropherograms) are shown in a list in the right pane. Edit the basecalling as described above.
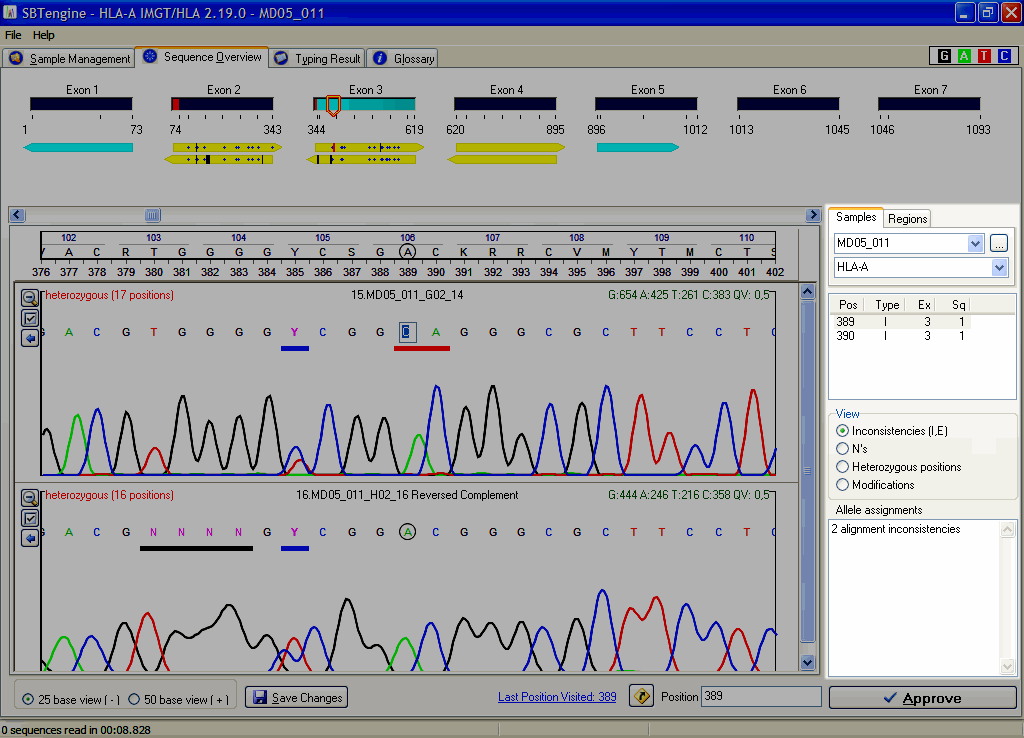
When you have resolved all inconsistencies, you will get a typing in most cases. SBTengine also shows the crucial positions in the right pane. You can distinguish the crucial postions list from the inconsistencies list by the light yellow background and radio button below which is set to "Crucial Positions" instead of "Inconsistencies".
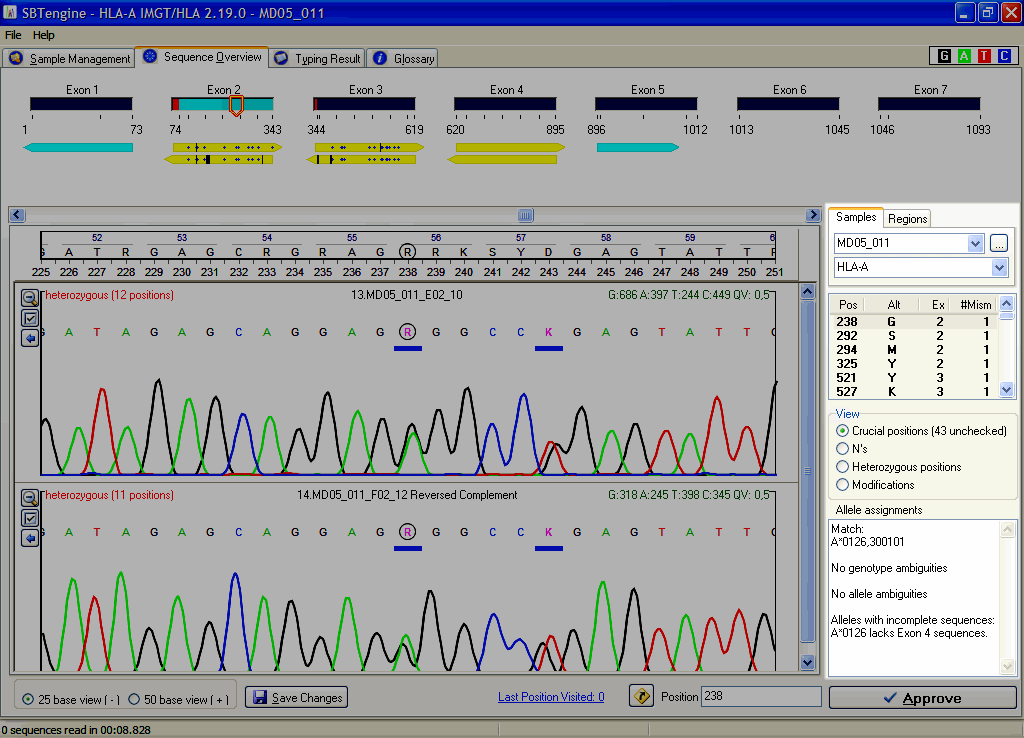
Step 3
Next, you have to check all crucial positions and verify if the basecalling is in concordance with the electropherogram. If required, the basecalling can be edited. SBTengine dynamically calculates the crucial positions and lists all relevant nucleotide positions, which, if changed, will lead to another allele assignment. Therefore it is crucial to check the basecalling at these positions. These positions can also be regarded as 'near matches'. When all crucial positions have been evaluated and all positions were clearly and correctly scored the allele assignment is ready.
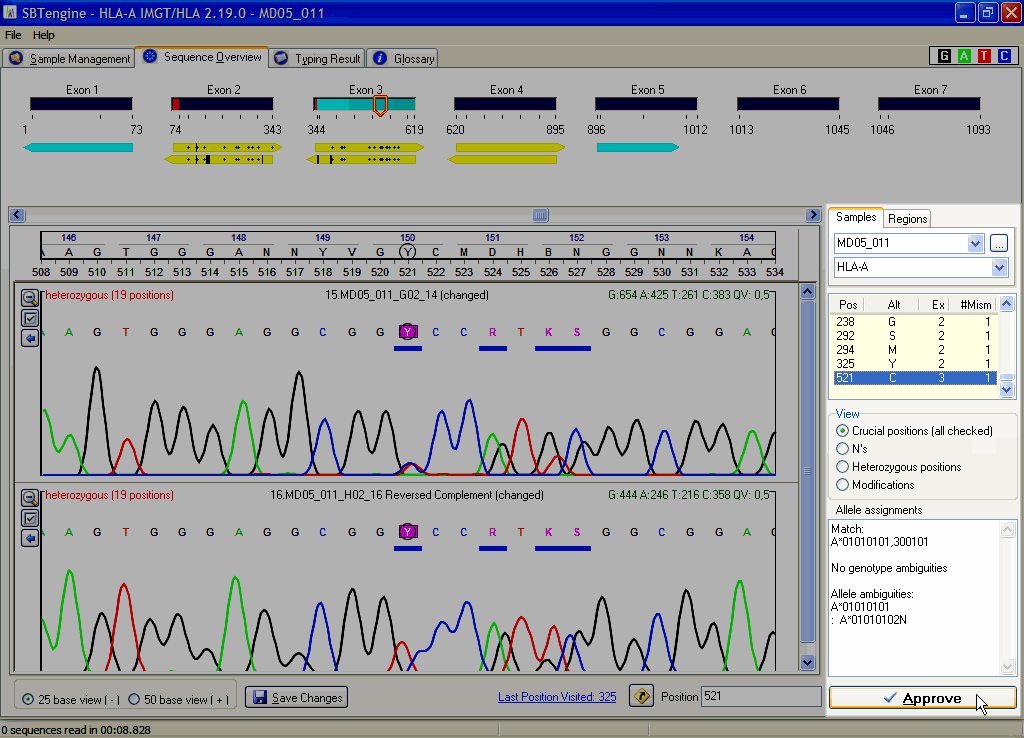
Finally, you need to approve the allele assignment by clicking the Approve button. SBTengine will ask you to provide your name to approve the typing. You can also enter a comment to the typing report, but this is not mandatory. Enter your name and press ENTER or click OK. The typing will be subsequently exported to an XML file.
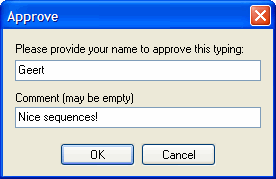
Alternatively, you may save the analysis first. All modifications are stored now. Another person (e.g. someone who has rights to authorize) may evaluate the analysis and approve consecutively.




