Contents
-
SBTengineŽ GPS
Introduction
SBTengine GPS is the newest tool in SBTengine that helps you to select GSSPs upfront and resolve ambiguities more efficiently.
In case pretyping information of a sample is available, it often is possible to determine which ambiguities can be expected and which GSSPs will resolve these ambiguities. Applying them in the first sequencing run will resolve the ambiguities, thus speeding up results and reducing analysis time.
Based on low-res pretyping information, GPS uses allele frequency tables to calculate the expected frequency of the sample being ambiguous after applying the core typing (exon 2 & 3). Then it calculates which GSSPs will resolve these ambiguities and to which extend.
How to use GPS
Select the GPS tab sheet in SBTengine
Select an allele frequency table
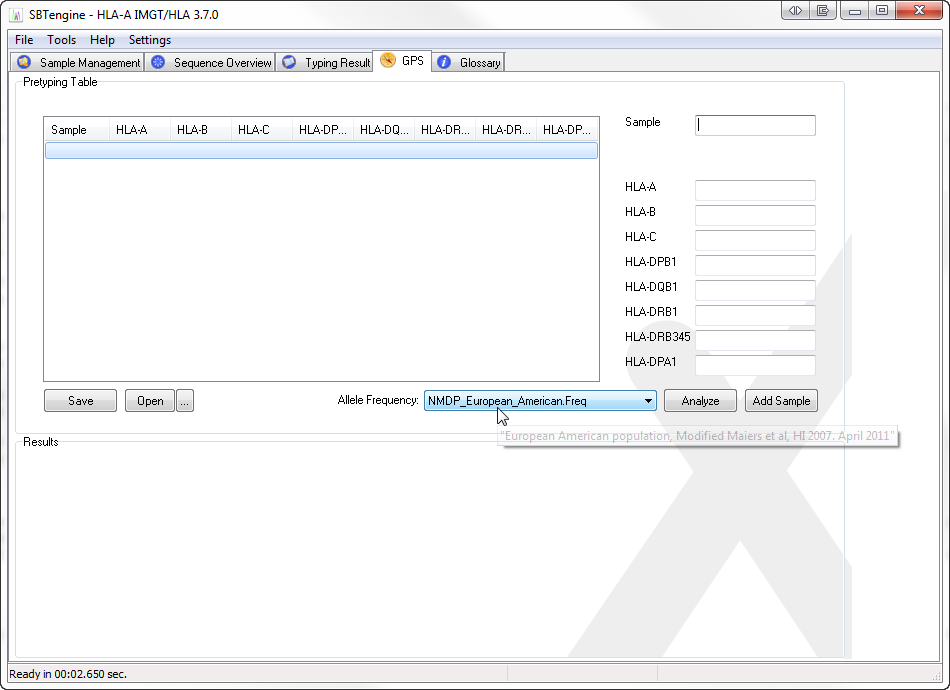
Add samples and their low-resolution or high-resolution typings
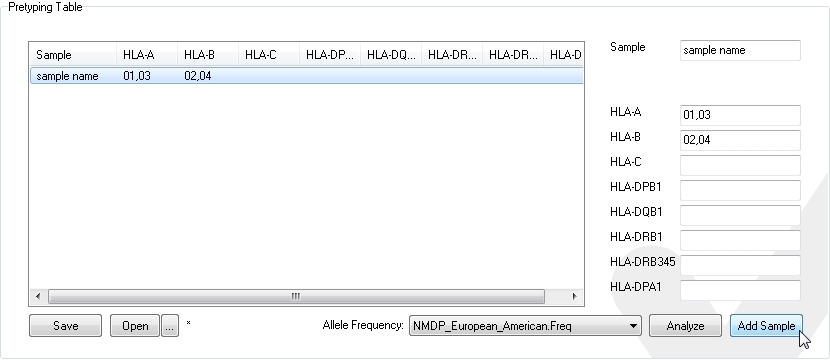
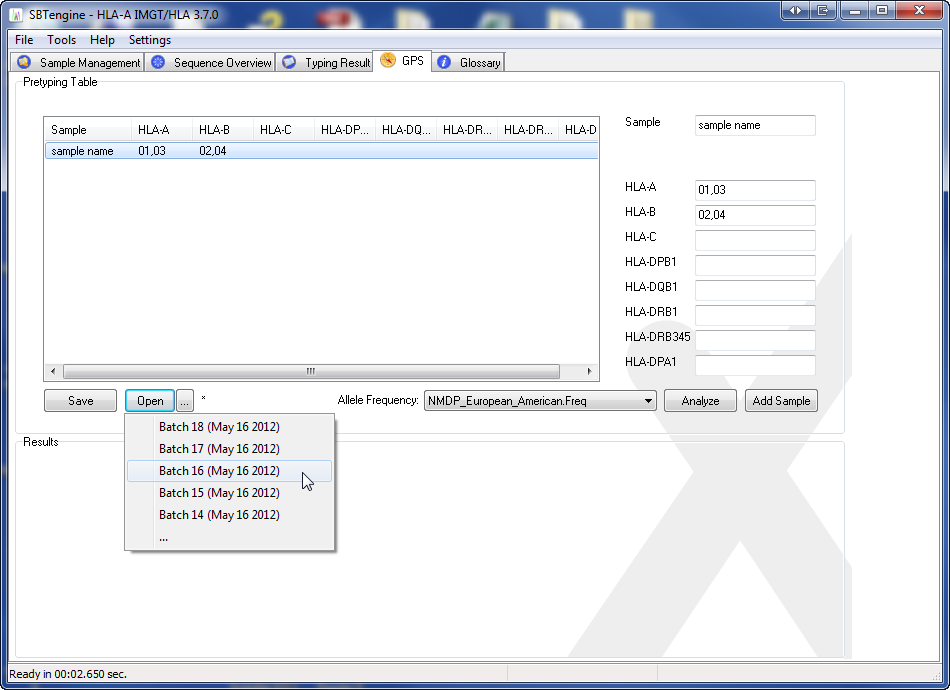
or load a pretyping table saved previously
Save the list of pretypings by clicking the save button.
Generate the list of resolving GSSP by clicking on the analyze button.
The list with recommended GSSPs is shown
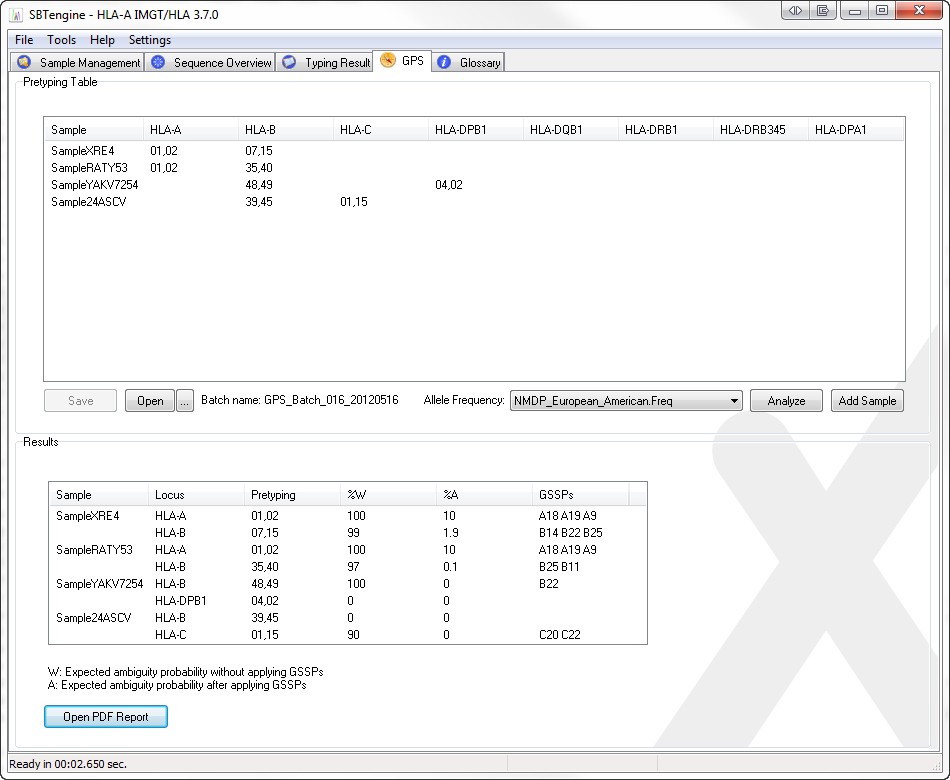
Click on 'Open PDF Report' to open the report, to enable printing.
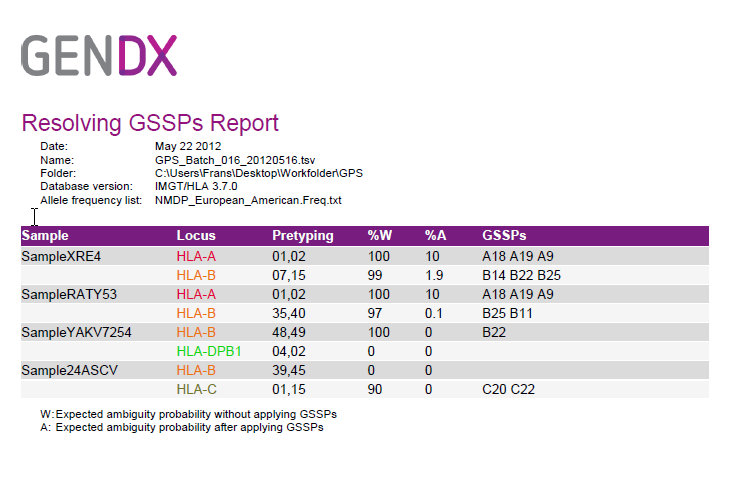
Applying the GSSPs recommended together with the core typing (exon 2 & 3) wil resolve the ambiguities.
The report contains 2 statistical numbers:
%W: The probability of the sample being ambiguous without applying any GSSPs
% A: The probability of the sample being ambiguous after applying the recommended GSSPs.
Since the GPS is based on allele frequencies, and on low-resolution pretyping data, the GSSPs recommended may differ from the GSSPs recommended by SBTengine after performing core typing. This is because the low resolution typing may also include ambiguities which will be resolved after core typing. In addition, GPS does not try to resolve between genotypes with very low frequencies, whereas SBTengine does.
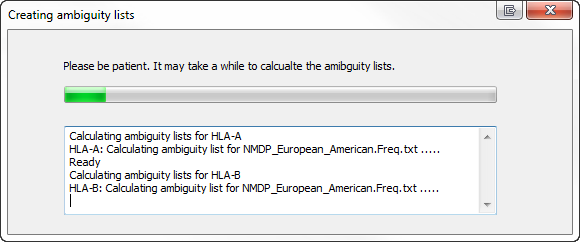
Creating ambiguity lists
GPS uses lists describing ambiguities. These lists will be generated from time to time. In general it will take some time to generate the ambiguity lists for all loci and all allele frequency tables. The lists will be saved, but will be regenerated with any change of IMGT library or allele frequency table.
To circumvent long waiting times, SBTengine has an option to generate these lists on demand, and thus can be generated after each update. This can be done by selecting in the menu 'Help' -> 'Calculate GPS data' -> 'All frequency lists'
Settings
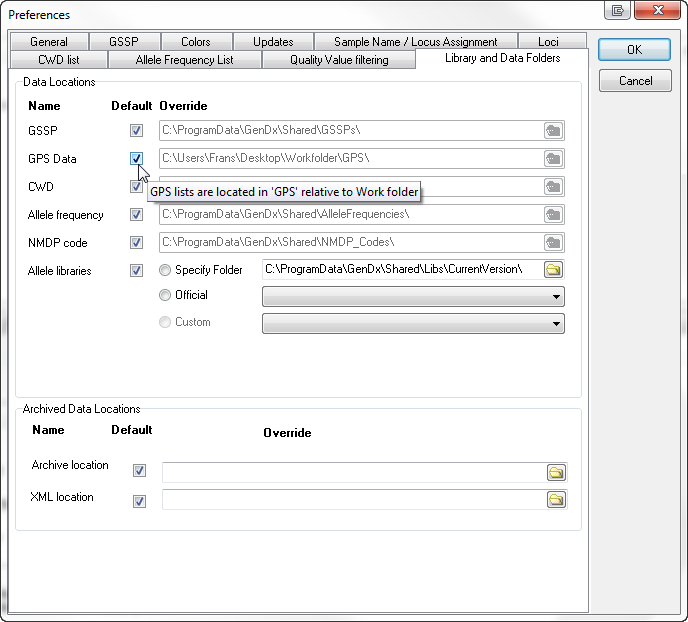
The GPS lists and the PDF files by default are saved in the 'GPS' folder located in the active workfolder. In the menu File, Preferences, tabsheet Library and Data Folders, the selection of GPS Data, the location can be changed.