Contents
-
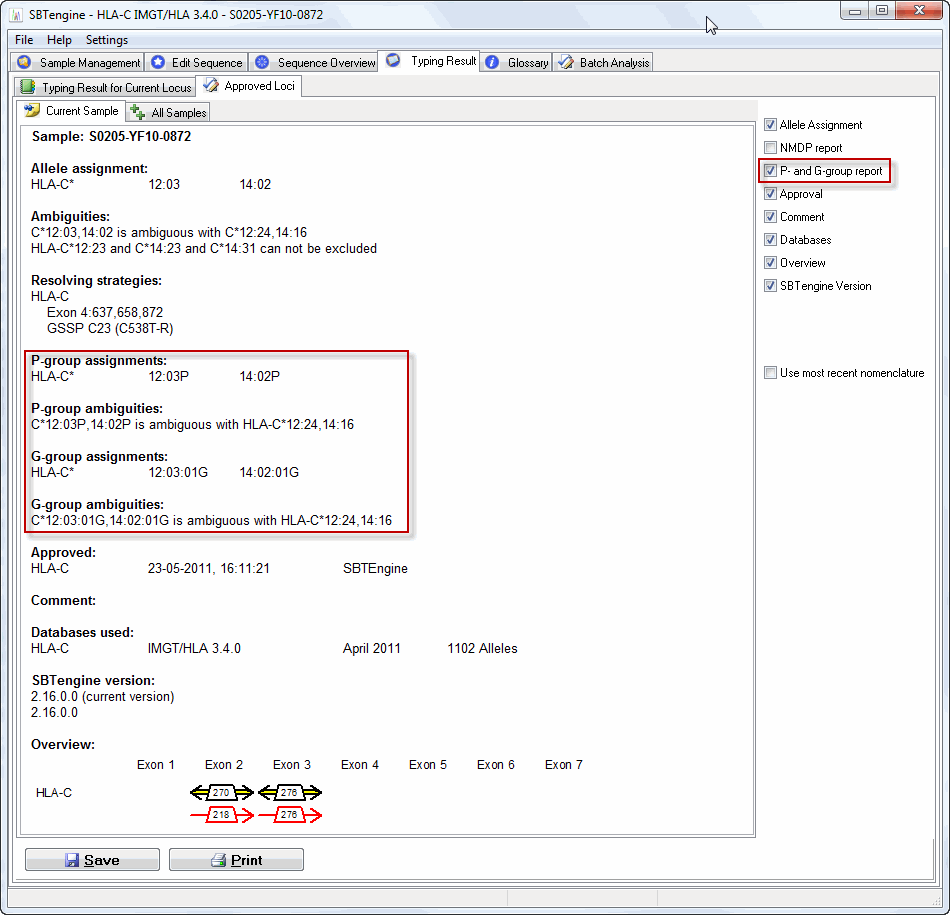
P and G Genotypes
P and G Codes are used to simplify the reporting of ambigious allele typings. These codes are defined in the HLA nomenclature as stated below (source http://hla.alleles.org).
G Codes For Reporting of Ambiguous Allele Typings
HLA alleles that have identical nucleotide sequences across the exons encoding the peptide binding domains (exon 2 and 3 for HLA class I and exon 2 only for HLA class II alleles), will be designated by an upper case 'G' which follows the first 3 fields of the allele designation of the lowest numbered allele in the group. The group designation will contain a minimum of six digits (source).
P Codes For Reporting of Ambiguous Allele Typings
This file contains details of all HLA Sequences having the same antigen binding domains. This analysis is performed on the protein sequence, and for HLA Class I alleles, identity in the 'antigen binding domains' is based on identical protein sequences as encoded by exons 2 and 3. For HLA Class II alleles this is based on identical protein sequences as encoded by exon 2. HLA alleles having nucleotide sequences that encode the same protein sequence for the peptide binding domains (exon 2 and 3 for HLA class I and exon 2 only for HLA class II alleles) will be designated by an upper case 'P' which follows the 2 field allele designation of the lowest numbered allele in the group. The group designation will contain a minimum of four digits (source).
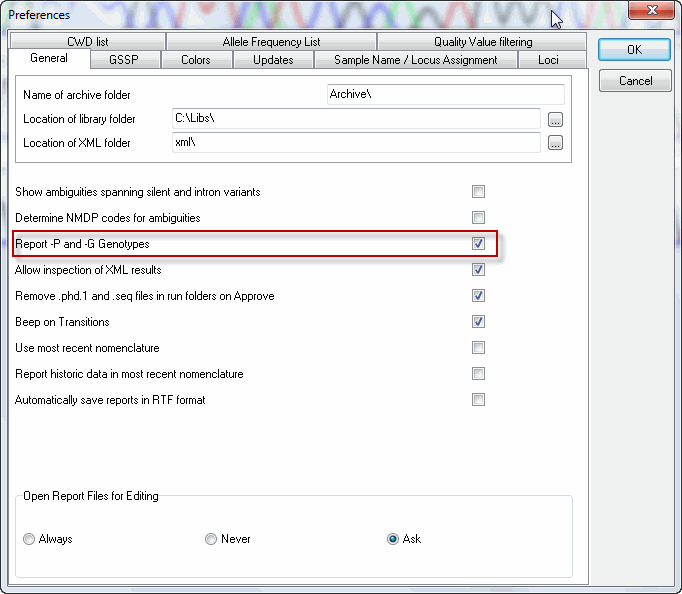
You can activate the reporting of P and G genotypes in SBTengine in the "File", "Preferences" menu by activating the check box "Report -P and -G genotypes" (see image below).
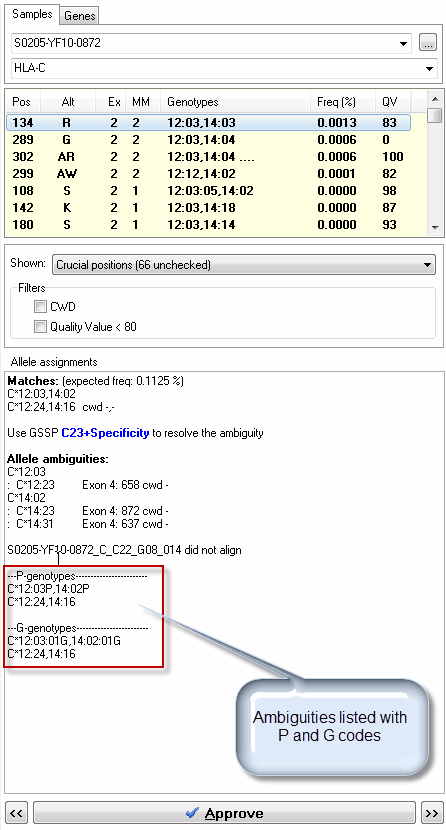
This will list the ambiguities in the "Allele Assignments" field also with P and G codes as shown in the image below.
In the typing reports, the P and G codes can also be shown by activating the check box "P- and G- group report". This is independent of the selection in the Preferences menu. So even if the function is toggled off in the Preferences menu, one can show P and G codes in the report (see image below). This information is also included in the XML files.