Insertions/Deletions
Insertions/Deletions
Insertions and deletions (in/dels) can be encountered in two ways in DNA sequencing based techniques.
1. The interpretation of the electropherograms by the basecaller software can be suboptimal when the individual peaks in the electropherogram are shifted. The distance between two peaks in a electropherogram should more or less be constant. When this is not the case, the basecaller has diffculties placing the basecalling at regular intervals. This can result in gaps (deletions) or additions (insertions) in the basecalling. These insertions/deletions are mostly limited to a few positions after which the basecalling becomes regular again. They can occur frequently in one sequence file. SBTengine indicates this type in/dels in the results pane of the Sequence Overview tab. The sequence file as well as the position is indicated. You can resolve the in/del by opening the particular sequence in the Edit tab, go to the indicated position and edit the basecalling accordingly thereby removing the in/del. By reopeneing the sequence in the Sequence overview, the in/del is resolved. Please note that the indicated postion of the in/del does refer to the position in the sequence i.e. in the Edit tab and not to the IMGT/HLA databse position i.e. in the Sequence Overview tab.
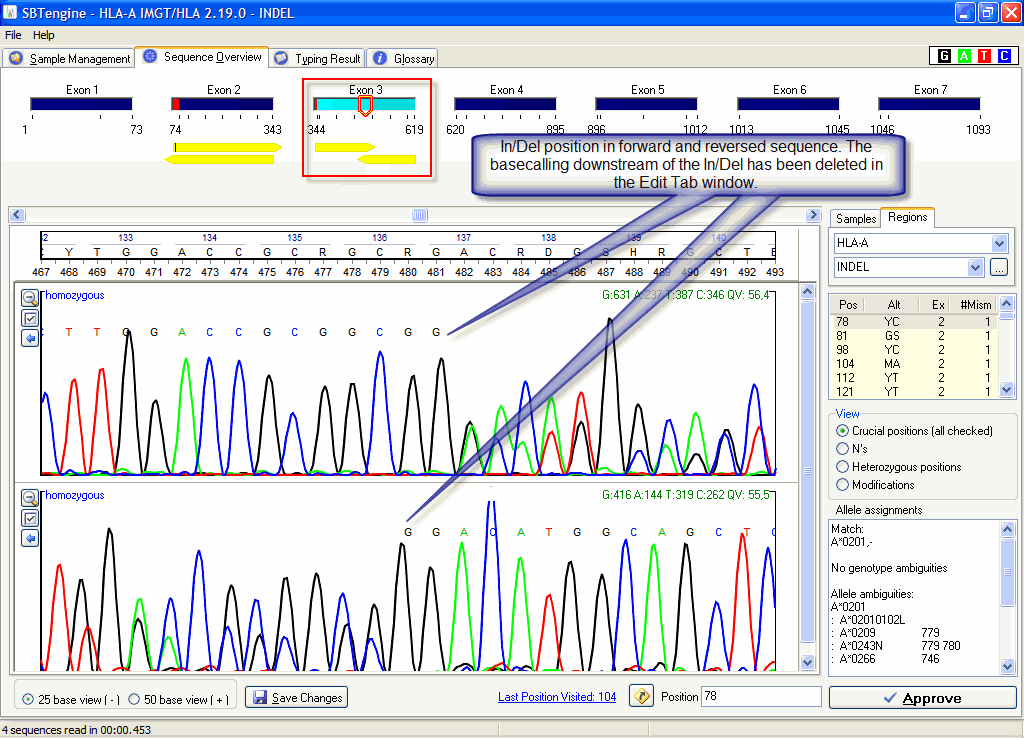
2. Insertions/deletions can occur when sequencing heterozygously i.e. both alleles of a sample are sequenced in one reaction. It does not matter whether the sample itself is homozygous or heterozygous. When one allele has a gap (deletion) or addition (insertion) in the DNA sequence compared to the other alllele the generated sequence will run out of frame from this point on, i.e. from this point on the generated sequence will only give heterozygous peaks and basecallings in the electropherogram. This hampers the alignment with the IMGT/HLA database when a sample is opened in SBTengine and the sequence will be rejected. When viewed in the Edit tab you can easily recognize these insertions/deletions as they typically resemble the picture below.

You may partly regain the sequence information in the Sequence Overview by deleting the stretches of heterozygous basecallings of the forward and reversed sequencing reaction and reopen the Sequence Overview. In this way you still have sequence information available although not completely in the forward as well as the reversed direction.