Graphical display of exons
Graphical display of exons
At the top of the sequence overview window you see the dynamic sequence overview. The bars indicating the exons of the HLA gene are shown in dark purple when the exon sequence information is available in the IMGT/HLA database. A grey bar indicates that in the current IMGT/HLA database no sequence information is available, whereas red regions indicate intron sequence information present in the database. The active exon is highlighted in blue. The start and end positions of the exons are shown below each bar and are numbered according to the IMGT/HLA database.
The orange magnifier indicates the actual position of the cursor in the electropherogram of the selected exon.
Below the bars, you can see the loaded sequence files aligned with the IMGT/HLA database exon bars. The sequence data are displayed as yellow arrows in the direction of the sequenced DNA-strand. In the picture above you can see a forward and reverse sequence for exons 2, 3 and 4. Besides you can see exon 1 read-throughs toggled off. In the yellow arrows, some symbols are depicted:
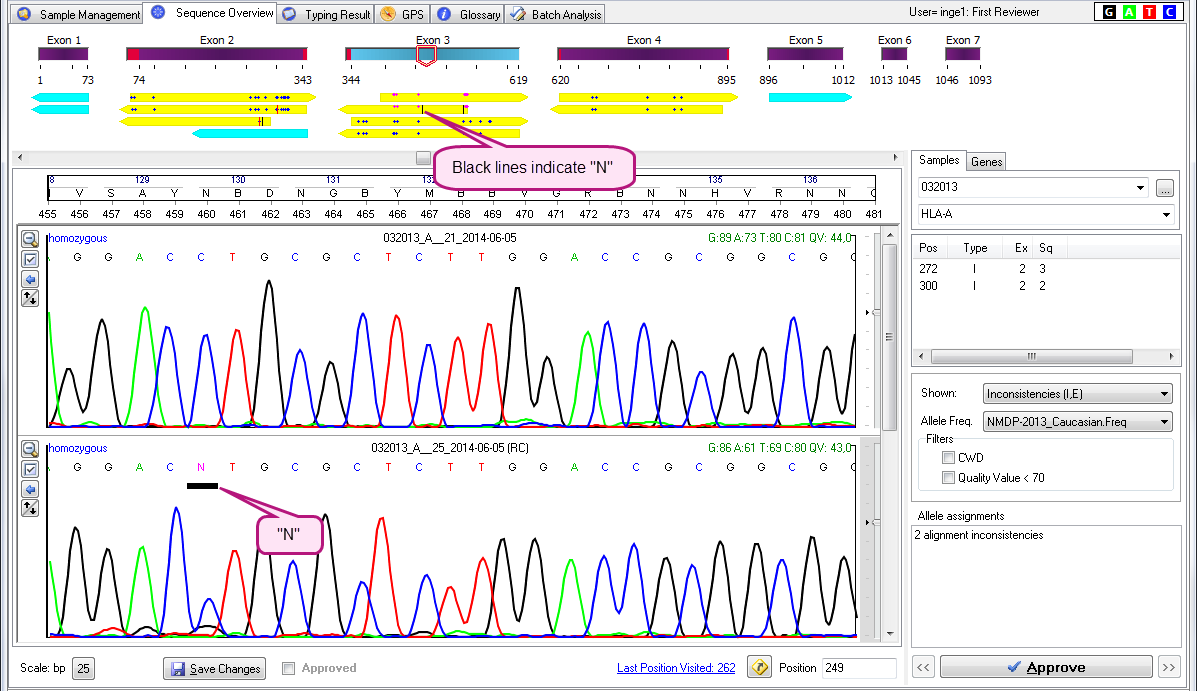
- Vertical black lines indicate "N" nucleotides.
- Vertical red lines show nucleotides that do not match the database (inconsistency).
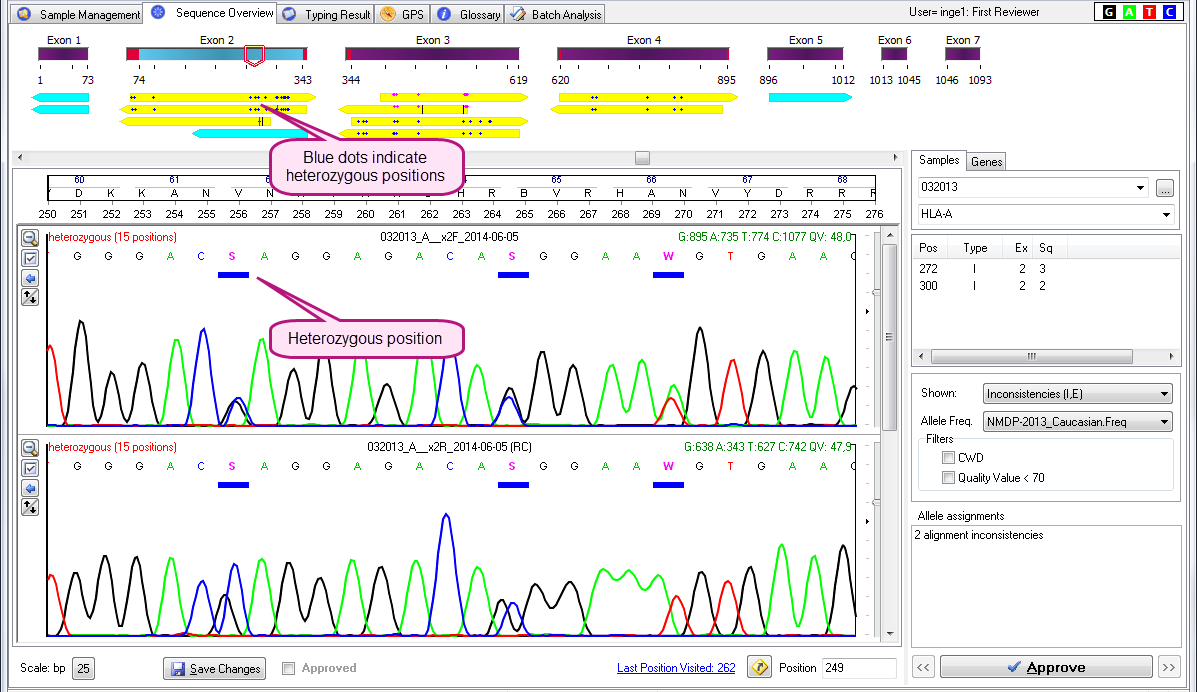
- Blue dots in the middle of the yellow arrow indicate heterozygous positions.
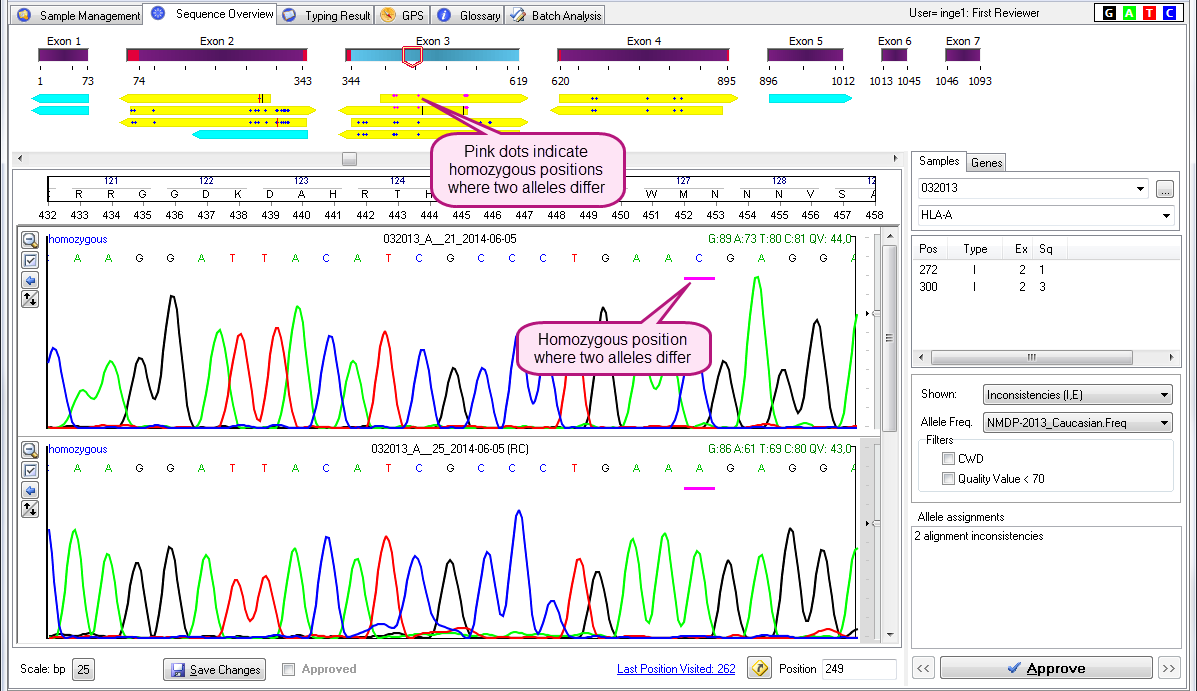
- Pink dots in the upper part of the yellow arrows indicate homozygous positions where the two alleles differ.
See the examples below:
Black lines
Red lines

Blue dots
Pink dots